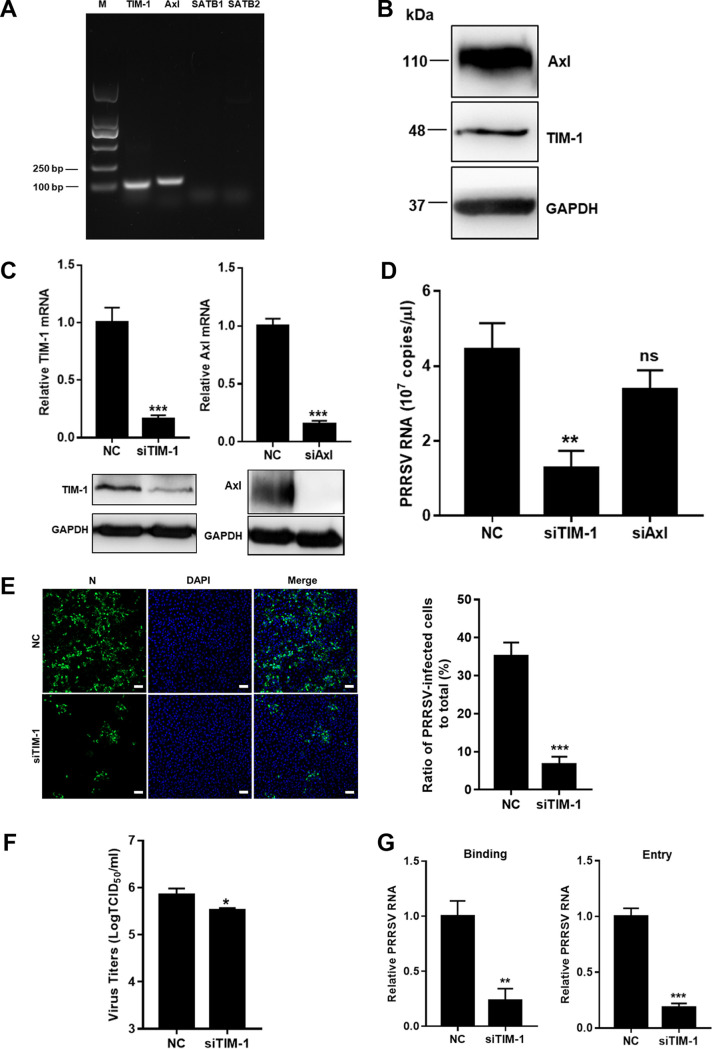
FIG 2.
TIM-1 is identified to recognize PRRSV as apoptotic mimicry in MARC-145 cells. (A) Transcription of PSRs in MARC-145 cells. MARC-145 cells were collected, and the reverse transcription cDNAs were prepared and subjected to PCR with the specific primers of TIM-1, Axl, Stabilin-1, and Stabilin-2. The PCR products of each gene fragment were subjected to agarose gel electrophoresis. (B) Expression of TIM-1 and Axl as determined by IB analysis. MARC-145 cells were harvested and lysed. TIM-1 and Axl were detected by IB. Knockdown of TIM-1 (C) significantly influenced PRRSV RNA abundance (D), infectivity (E), progeny viral titers (F), and viral binding (G). MARC-145 cells were transfected with siTIM-1, siAxl, or siRNA-NC for 36 h and infected with PPRSV (MOI = 10). The infected cells were collected for analyses of PRRSV RNA by RT-qPCR at 12 hpi, N protein expression by immunofluorescence at 24 hpi, viral titers by determining the TCID50 at 48 hpi, or binding and entry by RT-qPCR. Immunofluorescence images were quantified by counting the number of cells expressing viral N protein. Four random fields were counted per each condition, and the total number of cells per field was determined by DAPI staining. Each experiment was performed three times, and similar results were obtained. Differences between groups were assessed by using a Student t test, and the statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant). Scale bars, 50 μm.