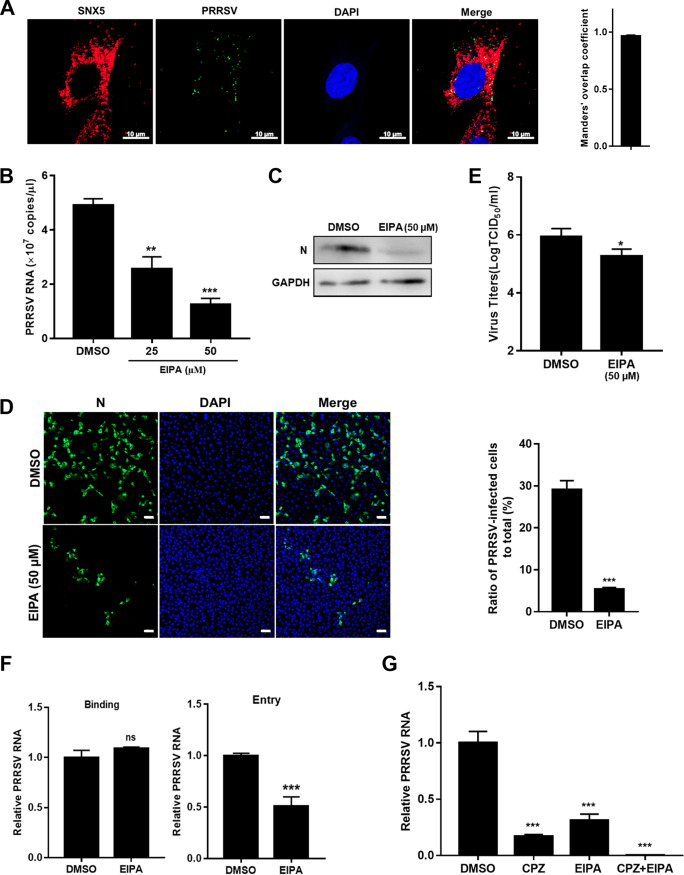
FIG 5.
PRRSV utilizes macropinocytosis to infect MARC-145 cells. (A) Colocalization of PRRSV and SNX5-marked macropinosomes. MARC-145 cells were inoculated with MOI = 10 PRRSV at 37°C for 30 min. Cells were fixed and stained with anti-PRRSV N protein (green) and anti-SNX5 (red) antibody. Nuclei were stained with DAPI. Confocal microscopy was performed to detect the location. The colocalization was assessed by determination of Manders’ overlap coefficient. Scale bars, 10 μm. The addition of EIPA decreased PRRSV RNA abundance (B), N protein expression (C), infectivity (D), progeny viral titers (E), and entry (F). The serum-starved MARC-145 cells were pretreated with 25 μM EIPA, 50 μM EIPA, or DMSO and infected with PRRSV BJ-4 (MOI = 10) for 1 h. The cells were collected for assessment of PRRSV RNA abundance by RT-qPCR at 12 hpi, N protein expression by IB or immunofluorescence at 24 hpi, viral titers using TCID50 at 48 hpi, or binding and entry by RT-qPCR. Scale bars, 50 μm. (G) Simultaneous addition of EIPA and CPZ almost abolished PRRSV infection. The serum-starved MARC-145 cells were pretreated with 50 μM EIPA and/or 10 μM CPZ and then infected with PRRSV BJ-4 (MOI = 10) for 1 h. The cells were collected for assessment of PRRSV RNA abundance by RT-qPCR at 12 hpi. Each experiment was performed three times, and similar results were obtained. Differences between groups were assessed by using a Student t test, and the statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).