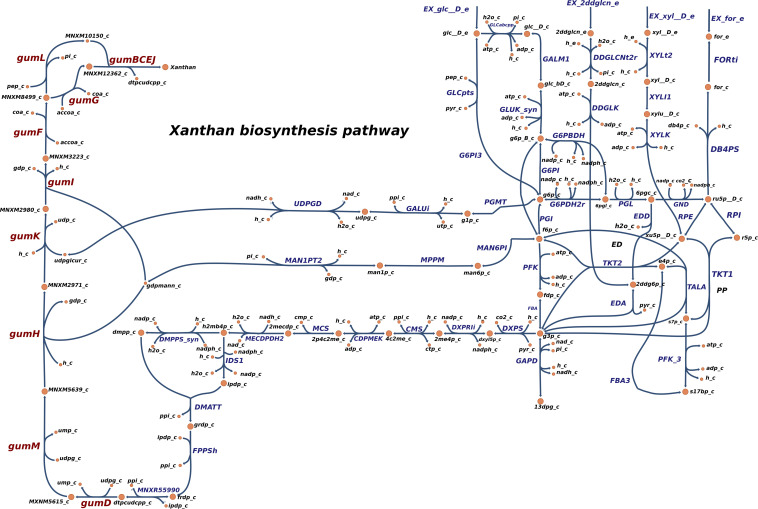
FIGURE 4.
Reconstructed metabolic map of xanthan biosynthesis in Xpm. Reactions belonging to xanthan biosynthesis pathway are shown to the left side: gumD,M,H,K,I,F,G,L and gumBCEJ. The carbohydrate utilization pathways are connected with the xanthan biosynthesis reaction by three branches (middle part of the graph). PGMT, phosphoglucomutase; GALUi, UTP-glucose-1-phosphate uridylyltransferase; UDPGD, UDPglucose 6-dehydrogenase; MAN6PI, mannose-6-phosphate isomerase; MPPM, D-mannose 6-phosphate 1,6-phosphomutase; MAN1PT2, mannose-1-phosphate guanylyltransferase; DXPS, 1-deoxy-D-xylulose 5-phosphate synthase; DXPRIi, 1-deoxy-D-xylulose reductoisomerase; CMS, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CDPMEK, 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase; MCS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; MECDPDH2, 2C-methyl-D-erythritol 2,4 cyclodiphosphate dehydratase; DMPPS, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; IDS1, isopentenyl-diphosphate synthase; DMATT, dimethylallyltranstransferase; FPPSh, farnesyl pyrophosphate synthase; MNXR55990, undecaprenyl diphosphate synthase. Carbohydrate utilization pathways: Entner–Doudoroff (ED), pentose-phosphate (PP), and Embden–Meyerhof–Parnas (EMP). Constructed in Escher (King et al., 2015).