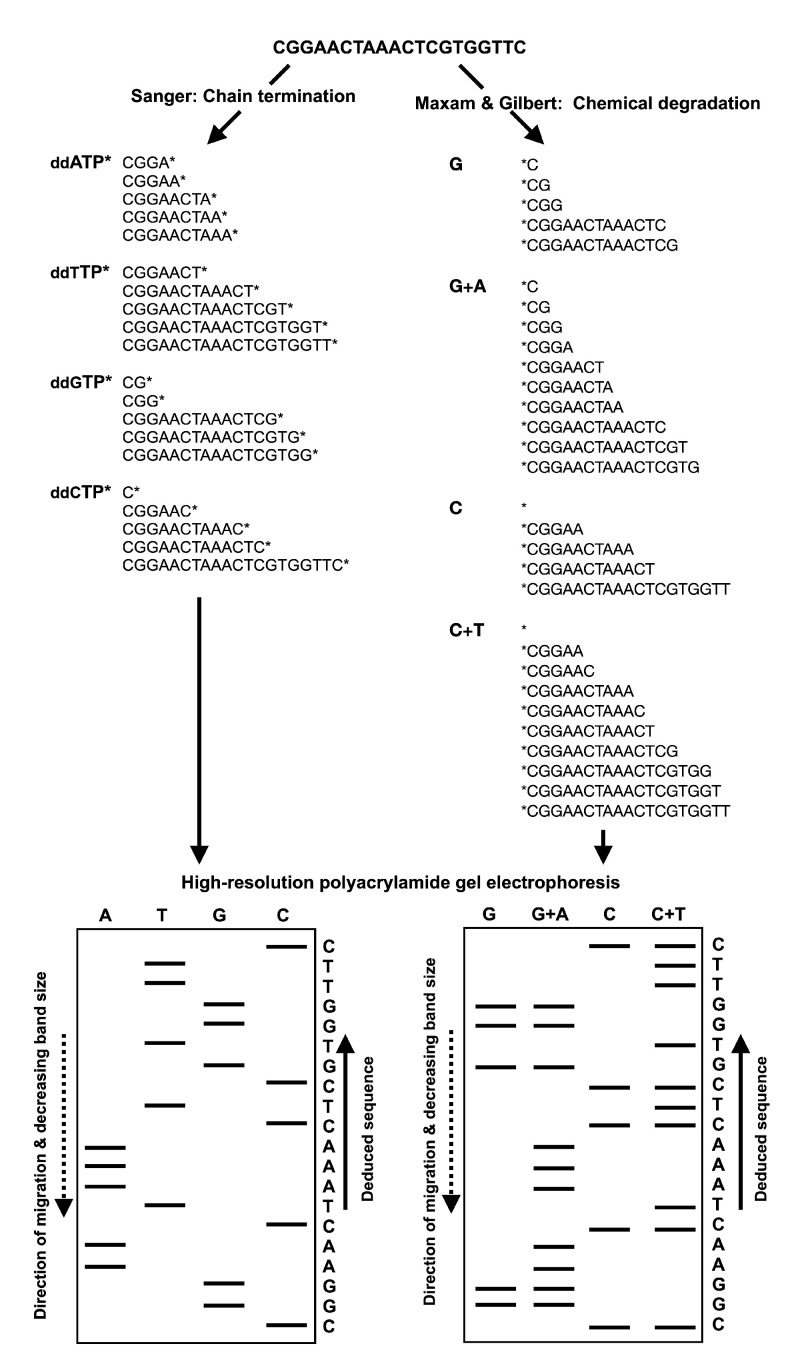
Figure 1.
Schematic illustrations of Sanger’s chain-termination and Maxam and Gilbert’s chemical sequencing techniques. In Sanger’s sequencing method, radiolabeled ddNTP nucleotides of a specific type (i.e., ddATP, ddTTP, ddGTP and ddCTP) are included in the DNA polymerase reactions at low concentrations along with the dNTPs. In each of the four reactions, polymerization will continue to extend with dNTPs until a ddNTP is incorporated, generating DNA strands of varying lengths. The DNA fragments are then visualized by high-resolution polyacrylamide gel electrophoresis. The nucleotide sequence is then deduced by finding the lane in which the band is present for a given site, as the 3′ terminating labeled ddNTP corresponds to the base at that position. Maxam and Gilbert’s method requires the radiolabeling (32P) of the 5′ phosphate moiety of the DNA fragment to be sequenced prior to chemical treatment for the selective removal of the base from a small proportion of the DNA molecules. Guanine is methylated by dimethyl sulfate, formic acid depurinates the purines (adenine and guanine); hydrazine hydrolyzes the pyrimidines (cytosine and thymine) and hydrazine in the presence of high salt (sodium chloride) concentrations can only react with cytosine. Piperidine is then used to cleave the phosphodiester backbone at the position of the modified base, yielding fragments of various lengths. The DNA fragments are then separated by high-resolution polyacrylamide gel electrophoresis in order to deduce the nucleotide sequence. The guanine (G) bands are present in both the G and A+G (purine) lanes, while the adenosine (A) band is present only in A+G lane. Similarly, cytosine (C) is indicated by the presence of bands in both the C and C+T (pyrimidines) lanes, while thymidine (T) bands are present only in C+T lane.