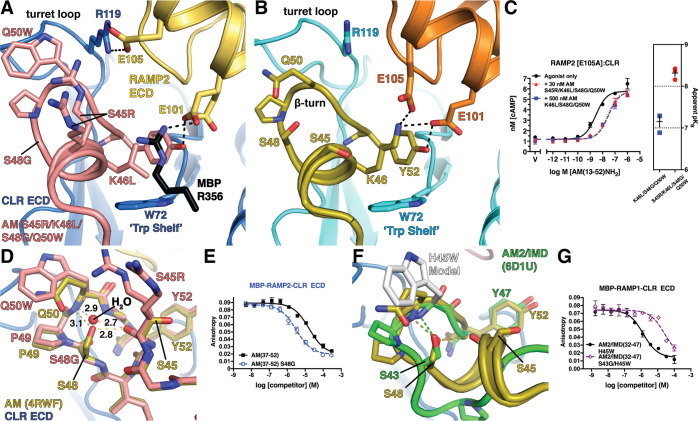
Figure 3.
Structural basis for enhanced RAMP2-CLR ECD affinity of AM(37–52) S45R/K46L/S48G/Q50W: (A) 1.83 Å resolution crystal structure of the variant bound to MBP-RAMP2-CLR ECD. (B) Crystal structure of AM-bound MBP-RAMP2-CLR ECD [PDB 4RWF] in the same view as panel A for comparison. (C) cAMP signaling antagonism assay in COS-7 cells using RAMP2 E105A:CLR and the indicated concentration of antagonist peptide variants. Right is a scatter plot of mean apparent pKB values determined from three independent experiments with error shown as SEM. (D) Superimposition of the new variant structure and 4RWF showing a detailed view of the peptide β-turn. H-bond distances are shown in angstroms. (E) Competition FP assay for the indicated AM peptides at MBP-RAMP2-CLR ECD. (F) Structural alignment of AM [PDB 4RWF] and AM2/IMD [PDB 6D1U]. (G) Competition FP assay for the indicated AM2/IMD peptides at MBP-RAMP1-CLR ECD. Mean pKI ± SEM values for the H45W and S43G/H45W variants were 6.08 ± 0.06 and 4.75 ± 0.08, respectively.