Figure 4.
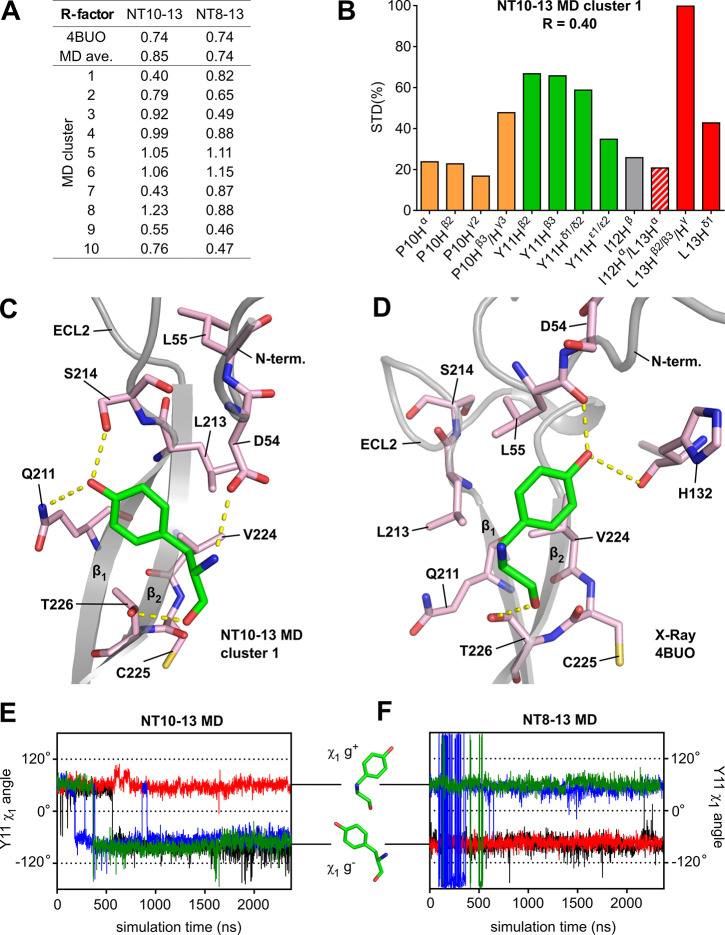
Alternative Y11 χ1 angle rotamer supported by epitope mapping. (A) R-factor ratios comparing epitope maps based on theoretical STD enhancements and the experimental STCDAF-CRL epitope map. Theoretical epitope maps were generated based on the NTS1 crystal structure (PDB 4BUO); based on the averaged STD enhancements over four MD trajectories (MDav) of NT10-13 and NT8-13 bound to enNTS1model; and based on the individual cluster representatives (clusters 1–10) extracted from the NT10-13 and NT8-13 bound MD trajectories. (B) Epitope map based on STD enhancements of NT10-13 MD cluster representative 1 normalized to the maximum value. This cluster showed the lowest R-factor (R = 0.40) among the theoretical epitope maps generated. The corresponding values are P10 Hα, 22%; P10 Hβ2, 23%; P10 Hγ2, 17%; P10 Hβ2/γ3, 48%; Y11 Hβ2, 66%; Y11 Hβ3, 65%; Y11 Hδ1/δ2, 56%; Y11 Hε1/ε2, 40%; I12 Hβ, 26%; I12 Hα/L13 Hα, 21%; L13 Hβ2/β3/Hγ, 100%; and L13 Hδ1, 48%. (C) Close-up of the ECL2 region of NT10-13 MD cluster representative 1 showing that the Y11 side chain adopts a χ1 g– conformation. (D) Close-up of the ECL2 region of 4BUO showing the Y11 side chain in a χ1 g+ conformation. Close-ups have been reduced to residues interacting with Y11 for clarity (two-dimensional ligand maps showing all receptor residues interacting with NT10-13 given in Figure S12). (E and F) Rotamer χ1 angle of Y11 in four independent MD simulations of NT10-13 and NT8-13, respectively, bound to enNTS1model.