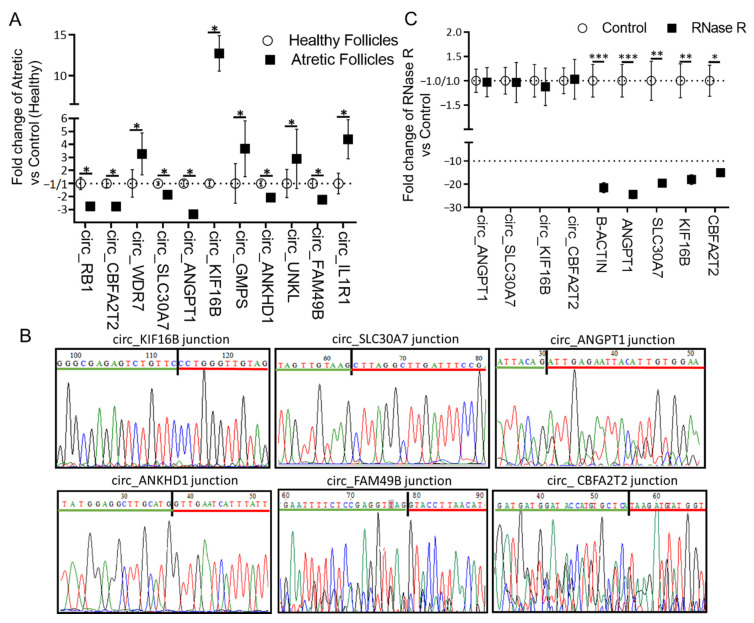
Figure 4.
Validation and characterization of circRNAs. (A) Relative circRNA expression measured by qRT-PCR. circRNA expression was calculated as a fold change of AA over HA follicles. circRNA expression in HA was set to 1 for up-regulated genes and −1 for down-regulated genes. (B) Representative examples of PCR products purified and sequenced to confirm circRNA junction sites by Sanger sequencing, n = 3. (C) qRT-PCR for the abundance of circRNAs (n = 6) and mRNAs (n = 6) treated with RNase R. The amount of circRNAs and mRNAs was calculated as fold change of RNase R over controls. The expression values in the controls were set to 1 for up-regulated genes and −1 for down-regulated genes. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Follicles were isolated in batches at three different time points, each batch containing 15–20 ovaries.