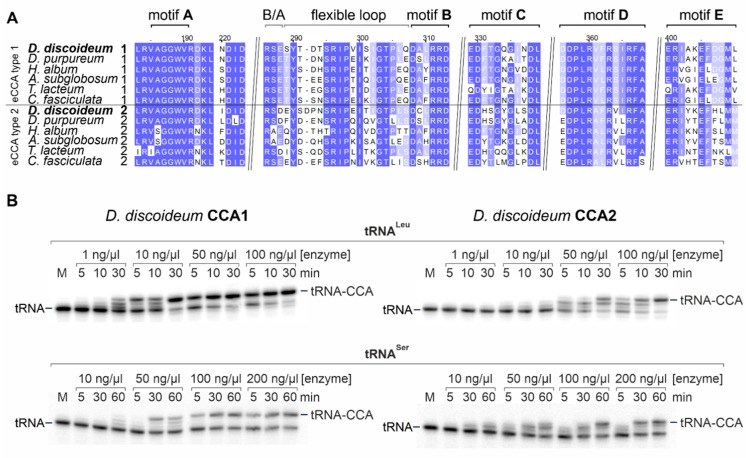
Figure 1.
Dictyostelia species possess two CCA-adding enzymes. (A) Alignment of the catalytic core motifs of tRNA nucleotidyltransferases in Dictyostelia species. All enzyme sequences carry the full set of motifs required for CCA addition. Yet, in an alignment of the complete protein sequences, the enzymes can be divided into subtypes CCA1 and CCA2 and each organism carries a pair of both enzyme types. Sequences are derived from NCBI (Accession numbers: D. discoideum 1: XP_629100.1; 2: Q55BE1; D. purpureum 1: XP_003291342; 2: XP_003283778; H. album 1: XP_020429327; 2: XP_020431701; A. subglobosum 1: XP_012758000; 2: XP_012749843; T. lacteum 1: KYQ91040; 2: KYQ90023; C. fasciculata 1: XP_004361592; 2: XP_004359300). (B) Recombinant CCA1 and CCA2 enzymes add a complete CCA-end to radioactively labeled transcripts of D. discoideum tRNALeu and tRNASer. The combined time and enzyme concentration series indicate that CCA1 is faster and more efficient in the reaction than CCA2. For both enzymes, tRNALeu seems to represent a better substrate than tRNASer, as it requires less enzyme concentration and incubation time to add a CCA-end, while for tRNASer, higher concentrations are required. Yet, both tRNA substrates are accepted by CCA1 as well as CCA2, indicating that the substrate itself does not affect the enzymes. M, mock incubation of labeled tRNA transcripts lacking the CCA-terminus; enzymes were added at indicated final concentrations. Incubation times are given in minutes (min).