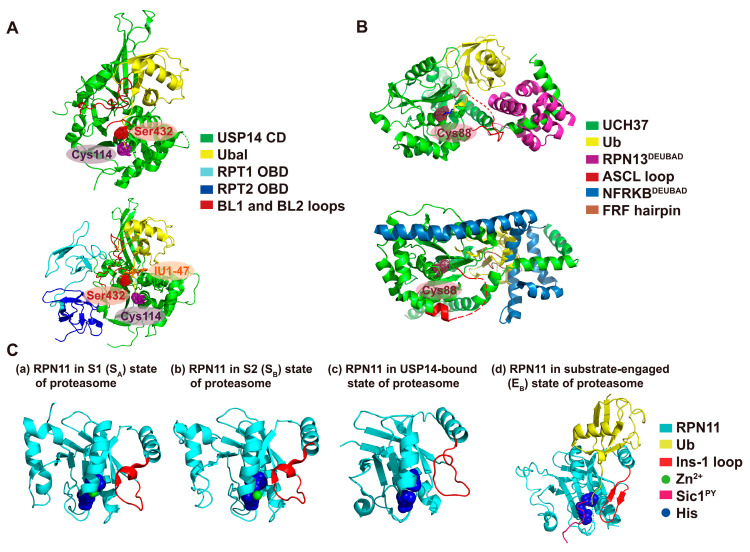
Figure 1.
Structures and conformational changes of proteasomal DUBs. (A) (Top panel) The crystal structure of the USP14 catalytic domain (CD) as a free form (PDB: 2AYN) [24]. Ubiquitin aldehyde (Ubal) is positioned on USP14’s catalytic domain as its inactive state to show steric hindrance of ubiquitin with BL1 and BL2 loops. The active site residues of USP14, including the catalytic Cys114 and conserved Ser432, are colored as stippled violet and red spheres, respectively. (Bottom panel) The structure of USP14-Ubal in human 26S proteasome complex that is resolved by cryo-EM (PDB: 5GJQ) [26]. Only the interactions between USP14 and OB domains (OBDs) of RPT1 and RPT2 are shown. IU1-47 was positioned by superimposing the crystal structure of IU1-47-bound USP14 (PDB: 6IIL) [28]. IU1-47 is shown as an orange stick model. (B) (Top panel) Structure of UCH37 in complex with RPN13DEUBAD domain and ubiquitin (PDB: 4UEL) [29]. The catalytic Cys88 of UCH37 is colored as stippled raspberry spheres. The ASCL loop is not fully resolved in the crystal structure, so the unresolved portion of the loop is indicated as a dashed red line. (Bottom panel) Structure of UCH37 in complex with NFRKBDEUBAD domain (PDB:4UF5) [29]. Ubiquitin is modeled on this complex based on the structure of UCH37-RPN13DEUBAD-ubiquitin and shows the steric hindrance with NFRKBDEUBAD. (C) Structure and conformational changes of RPN11 in the context of dynamic conformational states of the proteasome. Panels (a)–(d) represent each conformation of RPN11 in S1 (PDB: 5T0G) [30], S2 (PDB: 5T0H) [30], USP14-Ubal-bound human proteasome (PDB: 5GJQ) [26] and substrate-engaged human 26S proteasome (PDB: 6MSE) [31], respectively. RPN11’s active-site histidines are shown as a blue sphere model. Color coding of proteins and regions is given in the key.