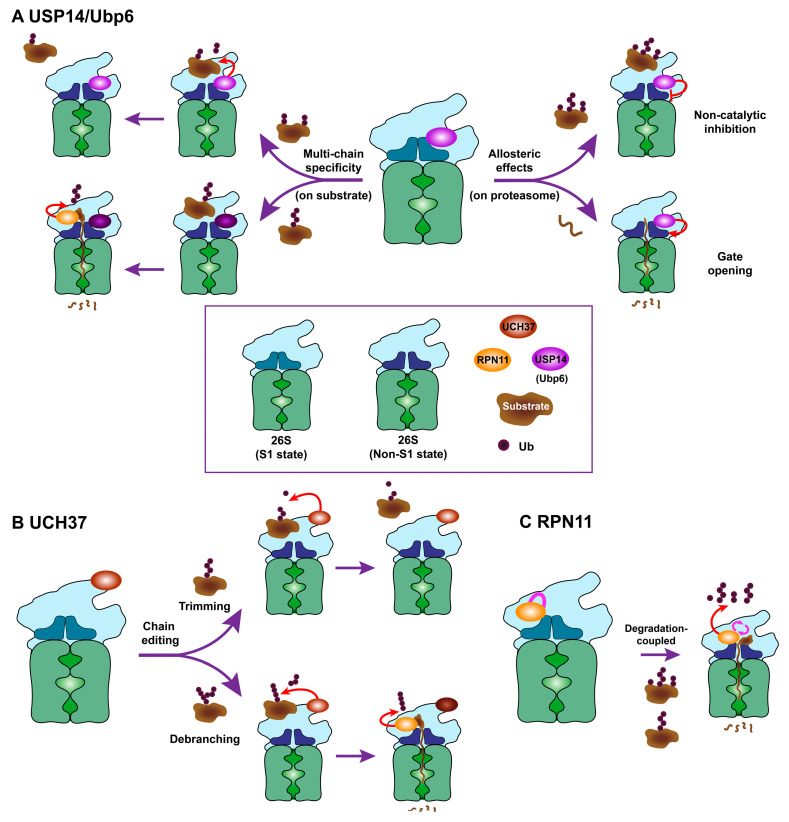
Figure 2.
Proposed working models for proteasomal DUBs. (A) (Left panel) Multi-chain specific and en bloc cleavage mechanism by USP14/Ubp6-mediated deubiquitinating activity. As shown in the upper panel, USP14/Ubp6 cleaves multi-chain ubiquitin conjugates at the proximal site. Depending on the characteristics of the remaining chain, the substrate deubiquitinated by USP14/Ubp6 can be spared from degradation prior to the commitment step. The lower panel indicates that a single chain bearing ubiquitin–protein conjugate serves as a poor USP14/Ubp6 substrate. This conjugate still can be subject to RPN11 activity, and subsequently undergoes degradation by the proteasome. (Right panel) USP14/Ubp6’s allosteric regulation on the proteasome. The upper panel shows that USP14/Ubp6 negatively regulates proteasome activity through its non-catalytic mechanism. In the lower panel, the model indicates that binding of USP14/Ubp6 to the proteasome in a certain conformational state (i.e., S2 state) induces the gate opening of the CP and enhances the uptake or degradation of the peptide substrate or some unstructured proteins. (B) Putative chain editing mechanism of UCH37 on the proteasome. In the upper panel, UCH37 acts by progressively trimming each mono-ubiquitin from the distal end of chains. This distal trimming may decrease the dwell time of substrates on the proteasome and thus rescue the substrate from degradation. The lower panel shows a recently reported debranching activity of UCH37 [14]. In contrast to the upper model, K48-linked branched chains can be selectively debranched by UCH37, resulting in promoted degradation by proteasome. (C) A typical working mechanism of degradation-coupled deubiquitination by RPN11. As ubiquitinated substrates are engaged and committed to degradation, RPN11 is shifted close to the substrate entry port of the ATPase ring. The Ins-1 loop of the active site also undergoes its conformational switch into a catalytically permissive β-hairpin structure, as depicted. Thus, RPN11 on the proteasome forms a functional module of deubiquitination activity, which is coupled to the unfolding–translocation–degradation axis. Where appropriate, DUB ovals that are darkly-colored inside indicate the inactive state of each enzyme.