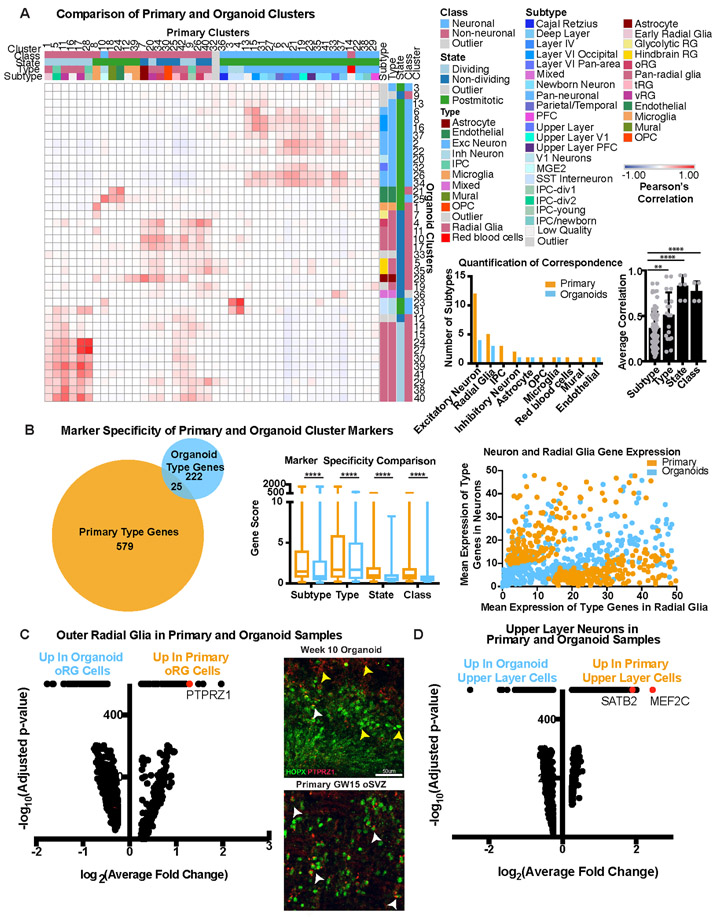
Figure 2. Molecular Comparisons of Cell Subtypes Between Primary and Organoid Samples.
a) Each cluster was classified by marker genes for class, state, type and subtype (primary: n=5 individuals across independent experiments; organoids: n=37 organoids from 4 PSC lines across 4 independent experiments). Correlation between pairwise combinations of marker genes in heatmap (red intensity: Pearson’s correlation from −1 to 1). First histogram indicates cell subtypes in primary (orange) and organoid (blue) samples. Second histogram shows quantitative correlation from the best match for each category averaged across clusters (mean plus standard deviation; subtype vs type: **p= 0.0073, subtype vs state: ****p=0.00008, subtype vs class: ****p= 0.003, Welch’s two-sided t-test). b) Using VariancePartition, the genes defining metadata properties were evaluated for contribution to overall variance. Genes contributing >25% variance by cell type were used in Venn diagram. Box and whisker (mean plus standard deviation) depicts level of specificity at class (****p=4.4e−14), state (****p=4.7e−18), type (****p= 1.02e−20) and subtype level (****p= 5.34e−38). For all comparisons Welch’s two-sided t-test was used (primary: n=5; organoids: n=37). The dot plot depicts discriminating genes between radial glia and neuron identity in primary samples. Each dot is a gene, shown as the average radial glia [x-axis] and neuron [y-axis] expression in primary (orange) or organoid (blue) cells. c) Differential expression (two-tailed Wilcoxon rank sum test) between clusters annotated as oRG cells in primary and organoid datasets generated log2(fold change) [x-axis] and −log10(adjusted p-value measurements) [y-axis] (primary: n=5; organoids: n=37). A pseudocount of 500 was assigned to comparisons with an adjusted p-value of 0. Many measurements were significant, including oRG identity gene, PTPRZ1. Week 8 organoids had minimal co-expression of PTPRZ1 and HOPX (top), while GW15 oSVZ contains extensive co-localization (repeated independently 3x). White arrows: double positive cells; yellow: single positive. Scale bar = 50 uM d) Differential expression (two-tailed Wilcoxon test) between cell clusters annotated as upper layer neurons.