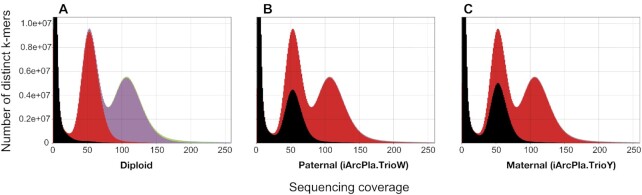
Figure 2:
k-mer spectra plots for the Arctia plantaginis trio-binned genome assembly. Plots produced using KAT, showing the frequency of k-mers in an assembly vs the frequency of k-mers (i.e., sequencing coverage) in the raw 10X Illumina reads, for the (A) combined diploid assembly (paternal plus maternal), (B) paternal-only assembly (iArcPla.TrioW), and (C) maternal-only assembly (iArcPla.TrioY). Colours represent k-mer copy number in the assembly: black k-mers are not represented (0 copies), red k-mers are represented once (1 copy), purple k-mers are represented twice (2 copies), and green k-mers are represented thrice (3 copies). The first peak corresponds to k-mers present in the raw reads but missing from the assembly due to sequencing errors, the second peak corresponds to k-mers from heterozygous regions, and the third peak corresponds to k-mers from homozygous regions. These plots show a complete and well-separated assembly of both haplotypes in the F1 offspring diploid genome.