Figure 3.
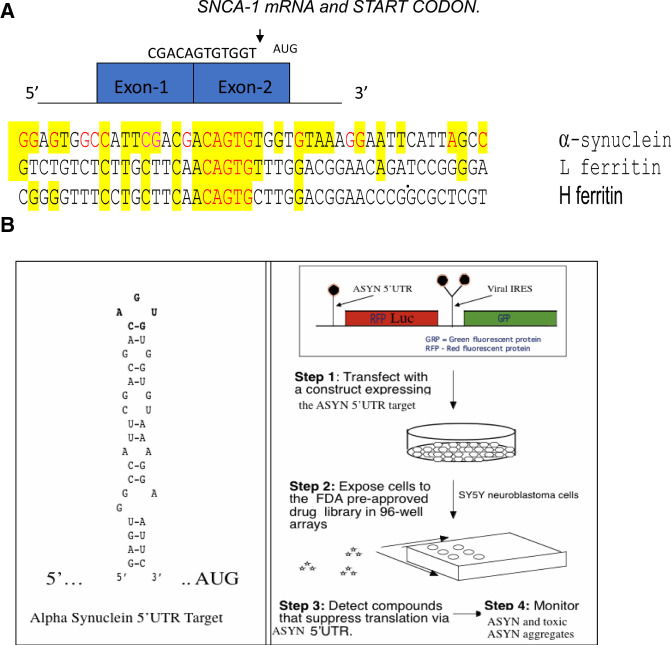
The 5′Untranslated region of alpha synuclein mRNA was the screening target for identifying αsyn translation blockers to address the need for Parkinson's disease and PDD and LBD therapies. The SNCA 5′UTR can be tested also to alleviate conditions of excess environmentally induced α-synucleinopathy (Friedlich et al. 2007; Rogers et al. 2011; Mikkilineni et al. 2012; Harischandra et al. 2019b). (A) The unique RNA target in the 5′UTR of the SNCA transcript. Depicted is the SNCA 5′UTR with its two exons immediately in front of the SNCA gene start codon (Olivares et al. 2009). Shown is the linear alignment of the SNCA 5′UTR with those for L- and H-ferritin mRNAs in the vicinity of their IRE RNA stem loops. (B) High-throughput screen for small molecule inhibitors of the RNA stem loop folded from the major neuronal neurodegenerative SNCA mRNA (MULTIFOLD, as described in Cho et al. 2010). Shown is a version of the transfection-based screen to identify SNCA 5′UTR directed translation inhibitors of a downstream luciferase reporter gene. During typical screens, inhibitors selectively maintained translation of a control GFP gene driven from an internal ribosome entry site (IRES) RNA structure. This RNA targeting technology was used to identify translation inhibitors of APP mRNA for AD therapy (Rogers et al. 2002a,b). The list of key HTS inhibitors of the SNCA 5′UTR are shown in PUBCHEM website as AID 1813. (Collaboration between Neurochemistry Laboratory at MGH and the Broad Inst. Cambridge, MA, https://pubchem.ncbi.nlm.nih.gov/bioassay/1813#section=Description