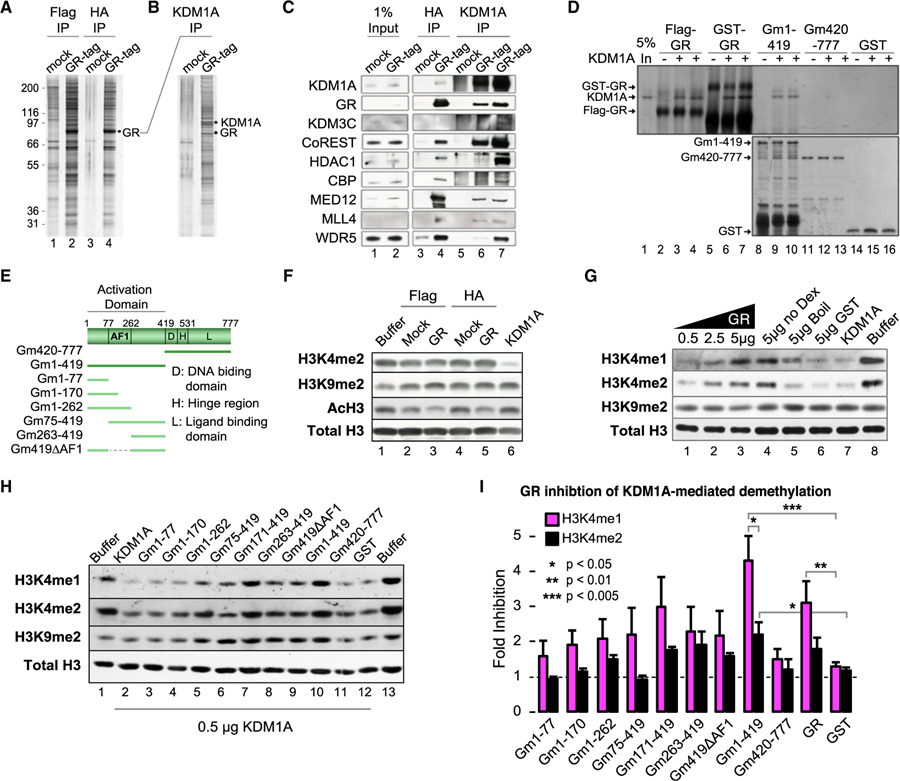
Figure 2. GR Forms a Complex with KDM1A and Modulates KDM1A H3K4-Specific Demethylase Activity.
(A) Silver stain of tandem affinity purified (TAP) GR complex and mock control. See also Figure S2A.
(B) Silver stain of anti-KDM1A reciprocal IP of mock or GR complexes.
(C) Western blot of GR complex components identified by tandem mass spectrometry (MS/MS). Lanes 3 and 4 are TAP samples (FLAG then HA purified). Lanes 5 and 6 are KDM1A IP from final TAP samples (underwent FLAG, then HA, then KDM1A IP). Lane 7 is KDM1A IP from the FLAG purification (underwent FLAG then KDM1A IP).
(D) Silver stain of KDM1A co-immunoprecipitation by GR. Purified full-length GR, either FLAG or GST tagged (lanes 2–7), GST-GR truncations, or GST control was used to pull-down KDM1A. Independent replicates loaded in adjacent wells. See also Figure S2B.
(E) Diagram of GR truncation mutants.
(F) Histone demethylase (HDM) assays with TAP complexes.
(G and H) Effect of full-length (G) and GR truncation (H) proteins on KDM1A histone demethylase activity. Recombinant GR protein was added to KDM1A HDM reactions and HDM activities were examined by immunoblot. To control for purification contaminants, a sample of purified GR protein was heat denatured before addition (G; lane 5). See also Figures S2C and S2D.
(I) Quantification of HDM assays by densitometry. Bars plot mean + SE of four to five biological replicates. p values by Student’s t test.