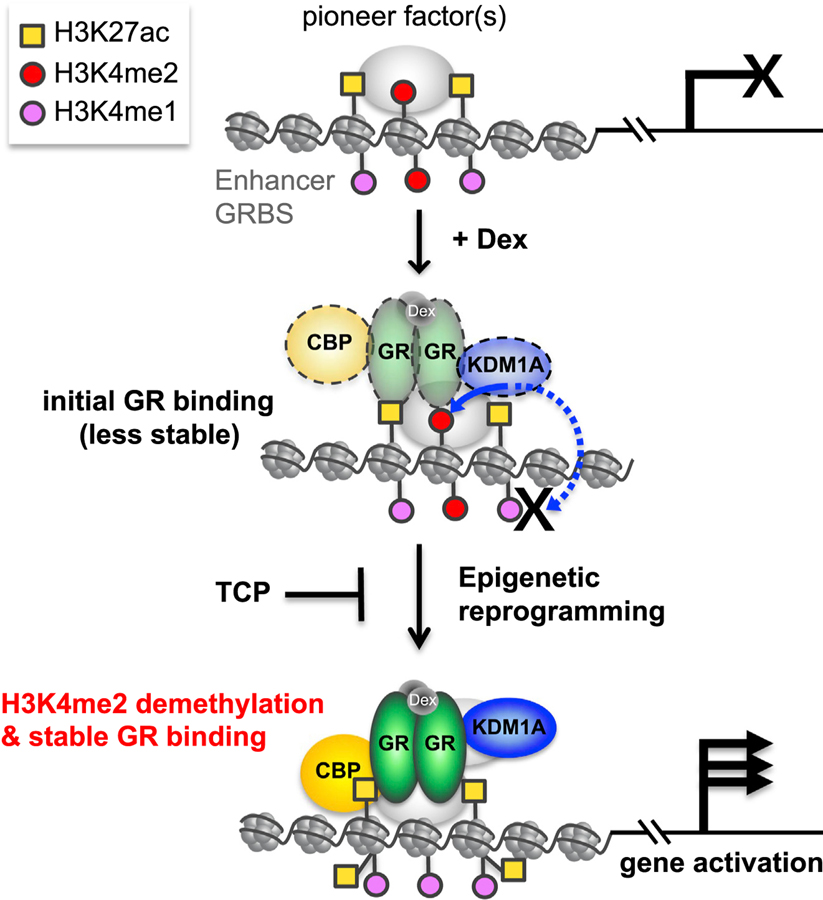
Figure 6. A Model of KDM1A-Mediated H3K4me2 Demethylation Contributing to GR-Mediated Gene Activation.
GRBSs, including enhancers potentially bound by pioneer factors, are premarked by H3K4me2 and other modifications, such as H3K27ac. Upon Dex activation, GR is targeted and transiently binds to the GRBS, where it recruits KDM1A and other cofactors such as CBP, for epigenetic reprogramming of the region. As an initial key step, KDM1A actively and preferentially demethylates H3K4me2 but leaves H3K4me1 intact due to inhibition from GR. H3K4me2 demethylation by KDM1A is required to stabilize GR binding. Other cofactors recruited by stabilized GR, such as CBP, increase H3K27ac at GRBSs, and this coordinated epigenetic reprogramming activates enhancers, thereby inducing GR-target gene expression, a process that can be blocked by the KDM1A inhibitors such as TCP.