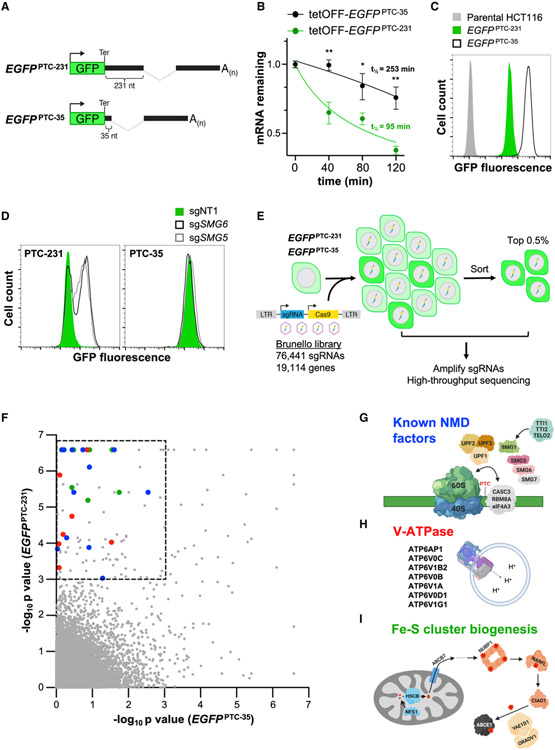
Figure 1. A Genome-wide CRISPR-Cas9 Screen Identifies NMD Regulators.
(A) Schematic of EGFPPTC-231 and EGFPPTC-35 reporter constructs. Ter, termination codon.
(B) Analysis of EGFPPTC-231 mRNA and EGFPPTC-35 mRNA stability in respective doxycycline-repressible (tetOFF)-EGFP cell lines. Cells were treated with 1 μg/mL doxycycline, and the EGFP mRNA level relative to GAPDH abundance at the indicated time points was determined by qRT-PCR.
(C) Flow cytometry analysis of EGFP fluorescence in the indicated cell lines.
(D) Flow cytometry analysis of EGFPPTC-231 cells and EGFPPTC-35 cells after lentiviral delivery of Cas9 and non-target (NT) or SMG5- or SMG6-targeting sgRNAs.
(E) Schematic of the CRISPR-Cas9 screening strategy.
(F) MAGeCK analysis of screening data. Hits within the dashed box represent those that met the significance threshold of p < 0.001 in EGFPPTC-231 cells and p > 0.001 in EGFPPTC-35 cells. Blue dots, known NMD factors; red dots, V-ATPase components; green dots, Fe-S biogenesis factors.
(G–I) Known NMD factors (G), V-ATPase subunits (H), and genes required for Fe-S cluster biogenesis (I) that were recovered as significant hits in the screen. Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, **p < 0.01; calculated by Student’s t test. See also Figure S1 and Table S1.