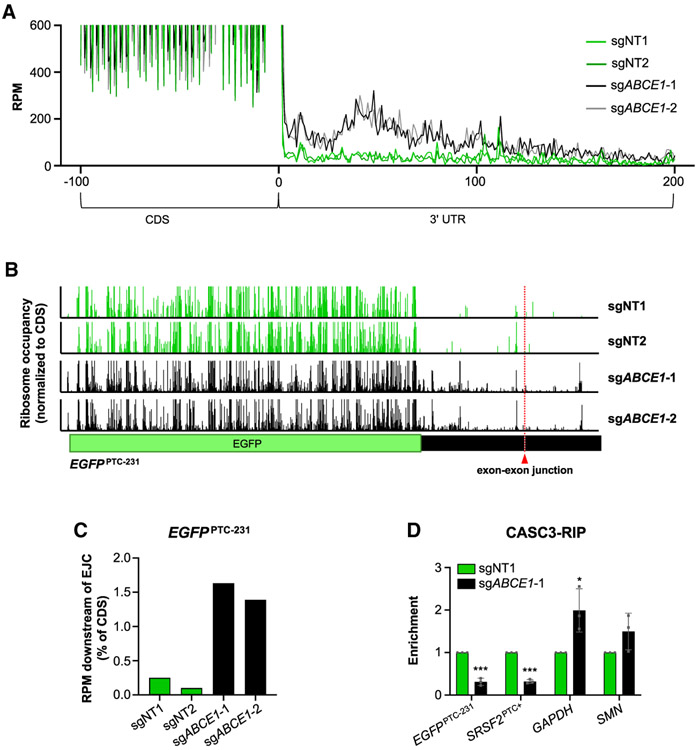
Figure 5. Unrecycled Ribosomes Downstream of Ters in ABCE1-Depleted Cells Displace EJCs.
(A) Metagene analysis demonstrating ribosome occupancy in 3′ UTRs. A window spanning 100 nt upstream of termination codon (position 0) to 200 nt downstream of termination codon is shown. RPM, ribosome-protected fragments per million mapped reads.
(B) Ribosome occupancy on the EGFPPTC-231 transcript after lentiviral delivery of Cas9 and the indicated sgRNAs. The position of the exon-exon junction in the in 3′ UTR of the reporter transcript is indicated. Ribosome occupancy is represented as the RPM value at a given position, normalized to the total RPM value in the EGFP ORF.
(C) Total ribosome occupancy downstream of the exon-exon junction in the 3′ UTR of the EGFPPTC-231 transcript after lentiviral delivery of Cas9 and the indicated sgRNAs. For each condition, the total RPM downstream of the exon-exon junction was normalized to the total RPM value in the EGFP ORF.
(D) qRT-PCR analysis of NMD targets (EGFPPTC-231 and SRSF2PTC+) andcontrol transcripts (GAPDH and SMN) in CASC3 RIP samples from cells expressing Cas9 and the indicated sgRNAs. For each transcript, enrichment was first normalized to input and then normalized to a paired control sample.
Data are represented as mean ± SD (n = 3 biological replicates for all qRT-PCR experiments). *p < 0.05, ***p < 0.001; calculated by Student’s t test. See also Figure S5.