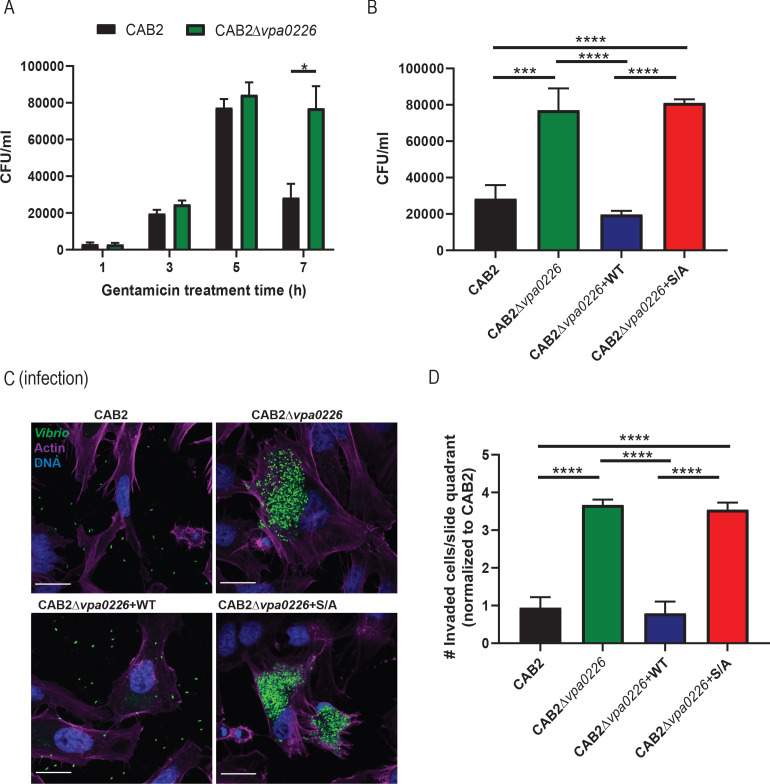
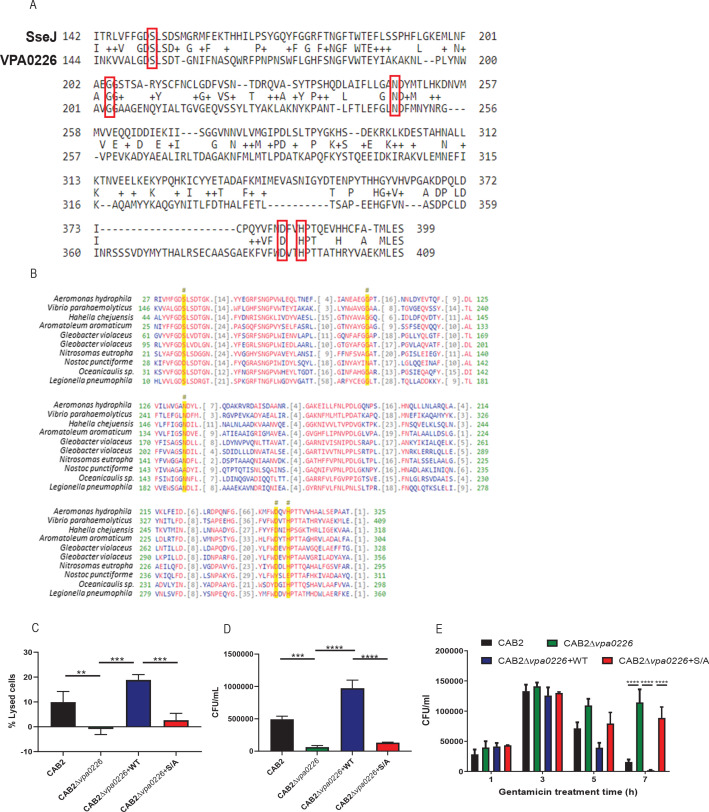
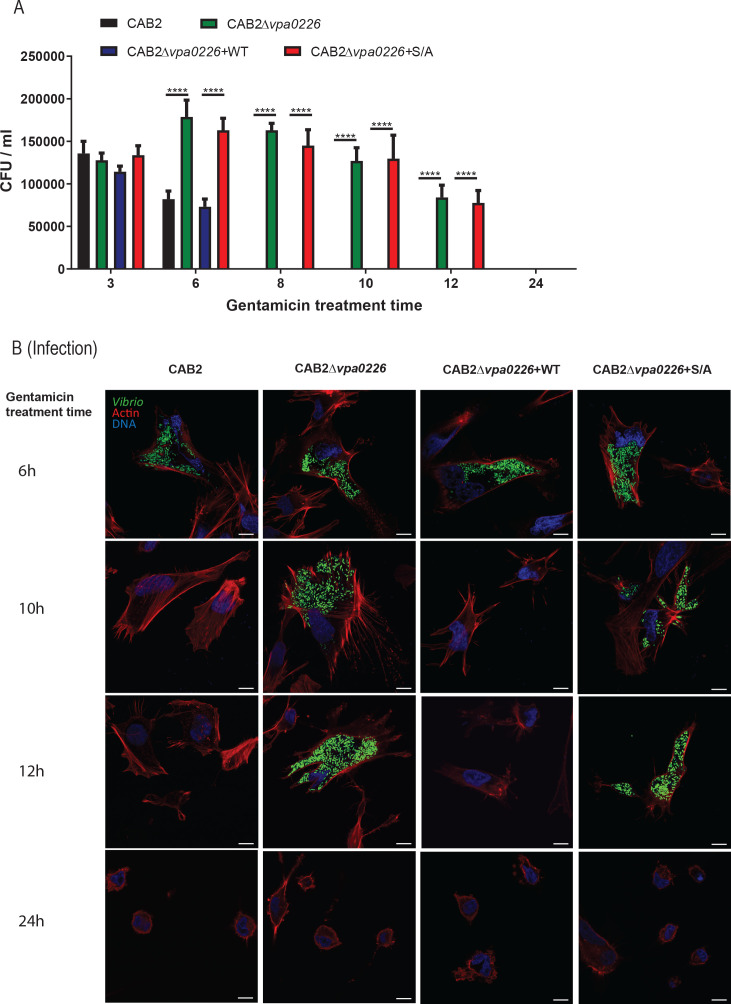
Figure 1. VPA0226 mediates bacterial egress from the host cell.
(A) HeLa cells were infected with CAB2 or CAB2Δvpa0226 for 1, 3, 5, and 7 hr PGT. Host cell lysates were serially diluted and plated onto MMM agar plates for intracellular bacterial colony counting (CFU/mL). Numbers are expressed as an average of three technical replicates for one of three independent experiments. Error bars represent standard deviation from the mean. Asterisks represent statistical significance (*=p < 0.05) using two-way ANOVA and Turkey’s multiple comparison test. (B) HeLa cells were infected with CAB2, CAB2Δvpa0226, CAB2Δvpa0226+WT, or CAB2Δvpa0226+S/A for 7 hr PGT. Host cell lysates were serially diluted and plated onto MMM agar plates for intracellular bacterial colony counting (CFU/mL). Numbers are expressed as an average of three technical replicates for one out of three independent experiments. Error bars represent standard deviation from the mean. Asterisks represent statistical significance (***=p < 0.001, ****=p < 0.0001) using one-way ANOVA and Turkey’s multiple comparison test. (C) Confocal micrographs of HeLa cells infected with GFP-expressing (green) CAB2, CAB2Δvpa0226, CAB2Δvpa0226+WT, or CAB2Δvpa0226+S/A for 7 hr PGT. Host cell actin was stained with Alexa 680-phalloidin (magenta) and DNA was stained with Hoechst (blue). Scale bars = 25 μm. (D, relative to C) Quantification of HeLa cells containing intracellular bacteria per quadrant of slide. Numbers were normalized to CAB2 and are expressed as an average of three independent experiments. Error bars represent standard deviation from the mean. Asterisks represent statistical significance (****=p < 0.0001) using one-way ANOVA and Turkey’s multiple comparison test.