Figure 1. Depletion of TRAIL promotes myoblast differentiation.
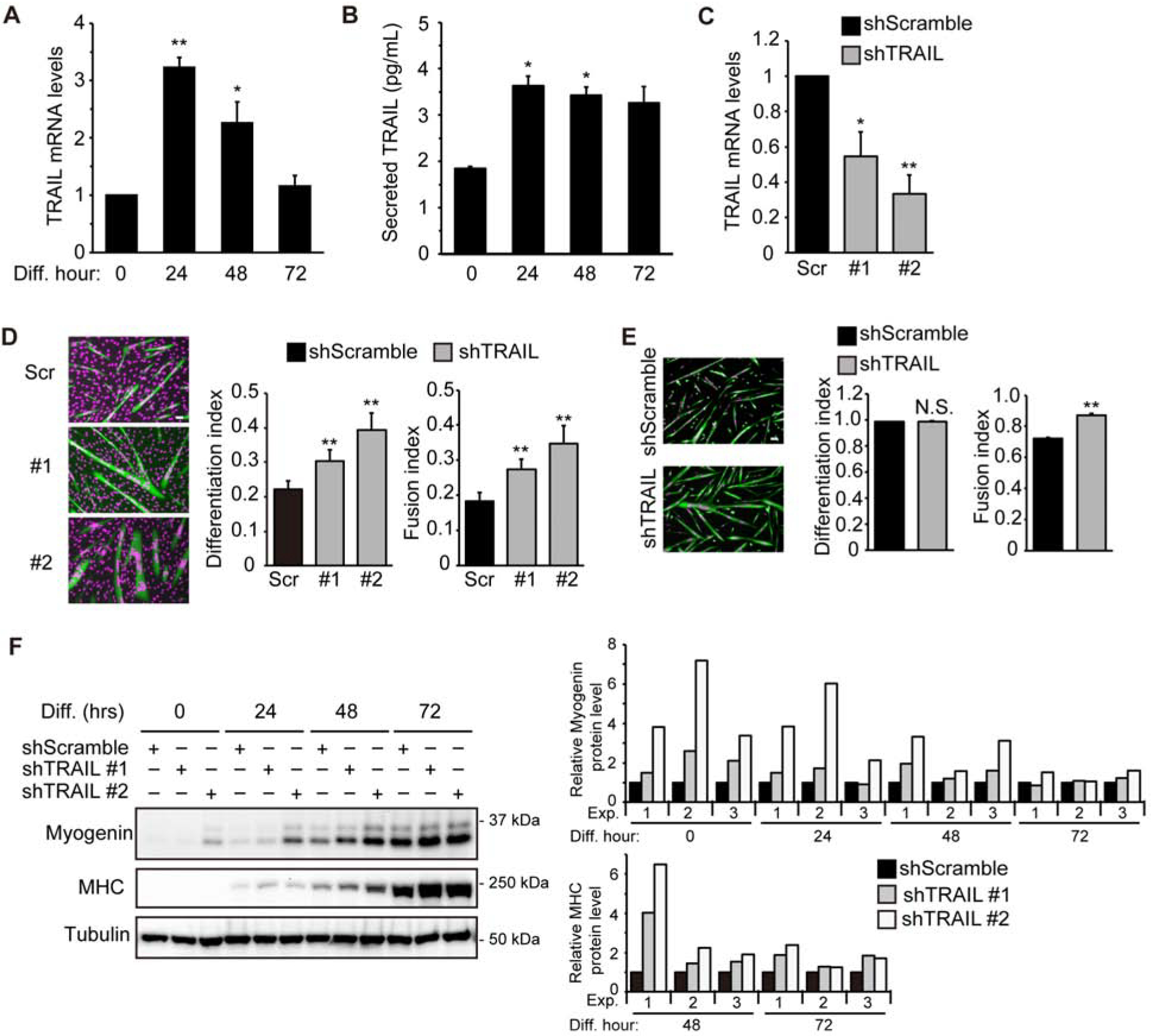
(A) RNA was isolated from differentiating C2C12 cells at different time points followed by qRT-PCR analysis (n=4). (B) Culture medium during C2C12 differentiation was collected at different time points and ELISA assay was performed to determine secreted TRAIL levels (n=3). (C) C2C12 cells were transduced with lentiviruses expressing shTRAIL or shScramble (Scr) followed by 2 days of puromycin selection. Cell lysates were then collected and subjected to qRT-PCR analysis (n=5). (D) Cells treated as in (C) were induced to differentiate for 72 hrs, followed by staining for MHC (green) and DAPI (magenta), and differentiation and fusion indices were quantified (n=7). (E) Mouse primary myoblasts were transduced with lentiviruses expressing shRNAs overnight, cultured in growth medium for 1 day, and then induced to differentiate for 48 hrs, followed by staining for MHC (green) and DAPI (magenta). Differentiation and fusion indices were quantified (n=3). (F) C2C12 cells treated as in (C) were induced to differentiate, and at different time points, cells were lysed and subjected to western analysis. Representative blots are shown. Results of quantification by densitometry are shown for 3 independent experiments. The MHC and myogenin protein levels were determined relative to the level of tubulin. Data were normalized to shScramble at each differentiation time point. When applicable, data are presented as mean with error bars representing SEM. Two-tailed t-test was performed to compare each data to control. *P < 0.05; **P < 0.01. NS, not significant. Scale bars: 50 μm.
