Abstract
Introduction
Adverse drug reactions (ADRs) are unintended reactions caused by a drug or combination of drugs taken by a patient. The current safety surveillance system relies on spontaneous reporting systems (SRSs) and more recently on observational health data; however, ADR detection may be delayed and lack geographic diversity. The broad scope of social media conversations, such as those on Twitter, can include health-related topics. Consequently, these data could be used to detect potentially novel ADRs with less latency. Although research regarding ADR detection using social media has made progress, findings are based on single information sources, and no study has yet integrated drug safety evidence from both an SRS and Twitter.
Objective
The aim of this study was to combine signals from an SRS and Twitter to facilitate the detection of safety signals and compare the performance of the combined system with signals generated by individual data sources.
Methods
We extracted potential drug–ADR posts from Twitter, used Monte Carlo expectation maximization to generate drug safety signals from both the US FDA Adverse Event Reporting System and posts from Twitter, and then integrated these signals using a Bayesian hierarchical model. The results from the integrated system and two individual sources were evaluated using a reference standard derived from drug labels. Area under the receiver operating characteristics curve (AUC) was computed to measure performance.
Results
We observed a significant improvement in the AUC of the combined system when comparing it with Twitter alone, and no improvement when comparing with the SRS alone. The AUCs ranged from 0.587 to 0.637 for the combined SRS and Twitter, from 0.525 to 0.534 for Twitter alone, and from 0.612 to 0.642 for the SRS alone. The results varied because different preprocessing procedures were applied to Twitter.
Conclusion
The accuracy of signal detection using social media can be improved by combining signals with those from SRSs. However, the combined system cannot achieve better AUC performance than data from FAERS alone, which may indicate that Twitter data are not ready to be integrated into a purely data-driven combination system.
Key Points
| This study is the first of its kind to use a computational method (empirical Bayesian model) to combine drug safety signals from a spontaneous reporting system with those from social media. |
| The accuracy of signal detection using social media can be improved by combining the signals with those from spontaneous reporting systems. |
| The evaluation of the combined system and individual sources was based on a fairly large reference standard, and the results of this study shed light on the potential role of Twitter data in pharmacovigilance. |
Introduction
Spontaneous reporting systems (SRSs) and a series of disproportionate analyses have been a cornerstone for pharmacovigilance [1]. However, this has many limitations, such as under-reporting, over-reporting of known ADRs, delayed reporting, and a lack of geographic diversity [2–4]. The rapid expansion and immediacy of social media websites such as Facebook and Twitter provides a broad coverage of health-related topics [5]. This means that these websites could be used to detect potentially novel adverse drug reactions (ADRs) with less latency [6]. A recent survey showed that about 3–4% of responding internet users had publicly shared their concerns about adverse reactions to medications on social media sites [7]. Regulators have become increasingly interested in mining such data from support group websites and social media postings as a potential new source for pharmacovigilance data [8, 9]. In 2013, the Association of the British Pharmaceutical Industry published guidance on the management of adverse events (AE) and product complaints sourced from digital media [10]. Although such guidance regarding the use of social media data for pharmacovigilance is lacking in the USA, the US FDA issued related regulations for publishing promotional material and risk/benefit information on social media [11].
The general pros and cons of using social media data for pharmacovigilance have been reviewed thoroughly [6, 12, 13]. One area of research focus is the application of natural language processing (NLP) and data mining to unstructured online sources with the aim of acquiring drug safety information. Notable among these is mining ADR signals from general purpose social networking sites such as Twitter [14] and from health support group websites such as PatientsLikeMe [15], DailyStrength [16], and MedHelp [17]. We chose to work with data from Twitter because of the large quantity of messages (> 500 million) distributed worldwide from a homogeneous source. A fundamental question was whether analysis of social media could lead to earlier detection of unknown AEs and therefore supplement SRSs. A further question was whether we could integrate analyses generated from social media and from an SRS to better detect ADR signals. Comparisons of these two types of data sources remain anecdotal and limited to the comparison of patient characteristics and reporting patterns [18, 19] or analyzing a specific task such as earlier detection by social media using a limited number (fewer than 15 pairs) of known positive and negative drug–ADR pairs [20], precluding any definite conclusions [21].
Previous studies have demonstrated that combining safety signals from several sources can improve the accuracy of signal detection. For example, augmented signal detection has been demonstrated by synthesizing signals generated from the FDA Adverse Event Reporting System (FAERS) and other individual data sources, including electronic health records (EHRs) [22, 23], claims data [23, 24], biomedical literature [25], chemical data [26], and internet search logs [12]. Recently, Harpaz et al. [23] developed multimodal methods to synthesize signals from four data sources: FAERS, claims data, the MEDLINE database, and the logs of major internet search engines. Piccinni et al. [27] developed a semantic web-based platform to integrate ADR resources from open data sources and social media. The integration of safety signals from social media with other data sources has not been studied.
The aim of this study was to systematically combine signals from FAERS and social media to facilitate the detection of safety signals. It is the first of its kind. Building on our previous Monte Carlo expectation maximization (MCEM) framework [28], we generated safety signals from each data source individually. We also pooled and aggregated signal scores from multiple data sources to produce composite signal scores, with an emphasis on more reliable data sources. We assessed the performance of this combined system together with signal detection based on the individual data sources using a retrospective evaluation method based on the reference standard of known side effects from drug labels.
Methods and Materials
Data Sources
Twitter Database
A collection of tweets over the 3 years from 2012 to 2014 were extracted from GNIP Decahose1, which provides a random sample of 10% of the real-time Twitter Firehose. A real-time sampling algorithm is used to randomly select the data. The initial collection involved approximately 50 billion tweets. We filtered out the re-tweets (33.5% of tweets) and non-English tweets (70.5% of the remaining tweets), yielding around 13 billion tweets.
FDA Adverse Event Reporting System (FAERS)
The FAERS data used in this study were pre-processed by Banda et al. [29]. This cleaned and standardized version of FAERS data involves the removal of duplicate case records and mapping of drug names to RxNorm concepts and ADR outcomes to Medical Dictionary for Regulatory Activities (MedDRA®) concepts. We used the same 3-year period for FAERS data as for the Twitter data, resulting in 2.3 million case reports.
Symptom Lists
A Twitter user may not use a professional medical term to describe a symptom. For example, “insomnia” may be described as “can’t sleep,” and “throwing up, chucking up, or puking” occur more often than “vomiting” in online social conversations. Therefore, a symptom dictionary that can map the symptoms in informal language to their appropriate professional medical terms is essential. The unified medical language system (UMLS) [30] incorporates moderately colloquial terminologies, such as the consumer health vocabulary (CHV) [31], which maps “throwing up” to “vomiting.” However, “chucking up” is not included. We also interrogated Wiki for another symptom list. We constructed three symptom lists to map the colloquial symptom-related terms to their professional terms using Wiki, UMLS1, and UMLS2. These three lists were used in our previous research and had reasonably broad coverage [14].
The first list of symptoms, named Wiki, was intended to capture symptoms expressed in layperson’s terms. This list was developed using the Wikipedia list of symptoms [32] in combination with those from previous work [33]. The list named UMLS1 involves terms from the UMLS semantic type T184 (sign or symptom). The list named UMLS2 extends the UMLS1 with additional semantic types involving T048 (mental or behavioral dysfunction) and T033 (finding). The UMLS1 and UMLS2 lists were generated using a local database installation of the UMLS. Table 1 shows statistics about these three symptom lists. Of these three, Wiki has the fewest symptoms but the highest number of synonyms per symptom, indicating that Wiki may include the most variants for a symptom. When comparing UMLS2 and UMLS1, adding T048 and T033 semantic types enlarged the number of symptoms almost 30 times, from 2733 to 68,720, but decreased the synonyms per symptom from 3.99 to 1.66.
Table 1.
Symptom lists statistics
| List | Symptoms | Unique synonyms | Synonyms/symptom |
|---|---|---|---|
| Wiki | 183 | 2016 | 11.74 |
| UMLS1 | 2733 | 8654 | 3.99 |
| UMLS2 | 68,720 | 105,721 | 1.66 |
UMLS Unified Medical Language System
Drug Lists
We started with drug names mentioned in the two data sources and used the RxNorm from the UMLS database to map these to their generic names. RxNorm provides normalized names for clinical drugs available in the USA and links the names to many of the drug vocabularies commonly used in pharmacy management and drug interaction software [34]. We expanded the list by adding known trade names that could be matched to generic names. The final list involves trade names as synonyms for their generic names.
Methodology Framework
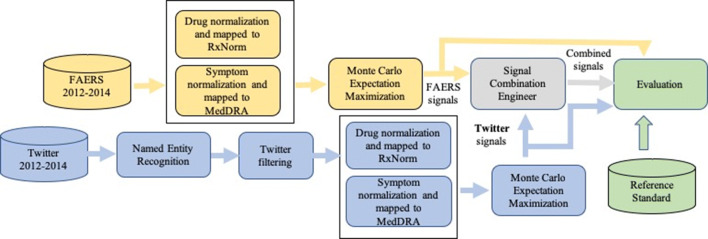
Figure 1 illustrates the proposed four-step pipeline for processing data and generating, combining, and evaluating ADR signals: (1) processing Twitter data by applying NLP and filtering methods to obtain structured coded data, (2) applying the MCEM method to generate signals from each data source, (3) combining signal scores from disparate databases with an empirical Bayesian approach, and (4) evaluating signal scores using a reference standard.
Fig. 1.
Processing pipeline for generating, combining and evaluating adverse drug reaction signals produced by Twitter, FAERS, and the combined system. FAERS US FDA Adverse Event Reporting System, MedDRA Medical Dictionary for Regulatory Activities
Processing Twitter Data
In our previous study, we manually annotated tweets using predefined named entities (NEs) from symptom lists and drug lists [35] and trained a linear chain conditional random field model [36]. The data set contains 1300 tweets with 253 mentions of diseases, 233 mentions of pharmacological substances, and 764 mentions of symptoms. The F1 performance of our system on this data set is 0.633 for diseases, 0.658 for pharmacological substances, and 0.679 for symptoms. The data set is available in our previous study (https://github.com/IBMMRL/medinfo2015).
We applied this model to identify tweets that mentioned relevant symptoms or drugs, resulting in approximately 230 million tweets. Note that most tweets were filtered out in this step. Furthermore, we used a mixed rule-based and machine learning pipeline to identify ADR-relevant tweets. First, we required tweets to mention drugs that appear in FAERS (18.9 million tweets, accounting for 8.4% of tweets from the last step). Second, we required a tweet to mention both a symptom (“disease” or “symptom”) and a drug (“pharmacological substance”) (553,000 tweets). Third, we developed a stop word list to remove mentions of erroneous drug names that we manually identified, such as “stay awake” (approximately 393,000 tweets). Fourth, we filtered out tweets that were advertisements, removing text that contained the token “http”, assuming that these were linked to advertisements, spam, or news articles. We also removed tweets containing the word “fact” since much of the spam used Twitter usernames such as “@AcneFacts”, “@thegoogleFact”, “@WhatTheFacts”, and “@FactBook”. Fifth, we removed tweets with drug terms that were too general, such as caffeine, cough syrup, vitamin D, zinc, and pain killer. After this step, 192,000 tweets were retained for the rest of our analysis. We previously tried to apply a machine learning method to filter out tweets that were indeed a treatment relationship between a drug and a symptom or disease rather than an ADR relationship, but the performance was quite poor [14], so we did not use this method in this study.
Generating Signals from FAERS Using Monte Carlo Expectation Maximization (MCEM)
MCEM is a modified version of the gamma Poisson shrinkage (GPS) model, with the aim to cope with the multidrug problem. The method assumes that each AE in each case report is caused by only one drug and then iteratively modifies the effective samples based on GPS signals (expectation step) and recalculates the GPS signals (maximization step) [28]. An independent comparison study showed that MCEM had the second highest area under the receiver operating characteristics (ROC) curve (AUC) and the highest Youden’s index [37] compared with other traditional disproportionality methods and performed very well in terms of high specificity based on its data set and evaluation strategy [38]. We used this method to generate signals from FAERS from 2012 to 2014.
Generating Signals from Twitter Data using MCEM
We considered each qualified Twitter post as a case report that could have been submitted to FAERS, so a single tweet that mentioned a drug name and an AE was a unit of analysis, and the overall qualified tweets were considered as an SRS. We mapped this tweeter-based SRS to a two-dimensional contingency table and used the MCEM to compute the associations, or signal scores. The time period was from 2012 to 2014.
Using an Empirical Bayesian Method to Synthesize Signals from Twitter and FAERS
We employed an empirical Bayesian strategy to combine drug safety signals obtained from FAERS and Twitter [28]. We cast the signal combination problem as a Bayesian hierarchical model that assumes signals from each source are independently and identically distributed with shared hyper-parameters. Mathematically, we indexed drug–ADR pair with , and as the quantified relationship between lth drug–ADR pair from kth () data source. In addition, we defined as the observed variance of . Then, the objective became to estimate the combined score for the lth drug–ADR pair with and . Here, we assumed the observed scores followed a Gaussian process centered around , where followed a Gaussian distribution centered around grand prior mean , which allows related signals to share statistical properties. These relations are given by the distributions defined in Eq. 1:
| 1 |
and the signal combination is computed as the estimate of as given by Eq. 2:
| 2 |
where is a summary statistic that is meant to summarize the signal scores provided by each data source for a given drug–ADR association. The summary statistic is for approximating the joint density of the scores and , which is used to obtain the posterior distribution of and . In addition, we denoted as the mean of the posterior distribution of given , and the scores.
In Eq. 1, we estimated and via expectation maximization with the independently distributed observations conditioned on . Thus, we could perform a maximum likelihood estimation of the hyper-parameters using the posterior distribution of given the scores and their variances in each iteration. Here, we defined such that the signal sources with less uncertainty would be emphasized more. To be specific, was calculated as a weighted average of the scores obtained by the same source first, then the average of the variances of individual scores was used as a weighting coefficient to combine different data sources. The formula is given in Eqs. 3 and 4.
| 3 |
| 4 |
where is the number of signals from the kth source.
Our signal combination step can be viewed as a pooling strategy. For the same drug–ADR pair, if the average uncertainty of one data source is high overall, then signal combination will have more weights on other data sources with less uncertainty. This approach also provides a smoothing effect: since each drug–ADR pair has safety scores from several sources, combining signals from multiple sources will prevent the performance of signal detection from degradation when there is artifact or data anomaly in one or more sources.
Evaluation
Reference Standard
To perform appropriate evaluation of the proposed system, we used a reference standard consisting of a set of positive controls (drug–ADR pairs known as true ADR relationships) and a set of negative controls (drug–medical condition pairs less likely to be associated). Several reference standards are used in pharmacovigilance, including the Observational Medical Outcomes Partnership (OMOP) reference standard [39], the EU-ADR reference standard [40], and a time-indexed reference standard [41]. However, most of the drug–ADR pairs in these reference standards are related to serious ADRs that are rarely mentioned in Twitter. Therefore, we chose to develop a reference standard based on Side Effect Resource (SIDER), a database that contains information on marketed medicines and their recorded ADRs [27] and has broader coverage for both drugs and ADRs, especially for mild ADRs. Its information is extracted from public documents and package inserts and is updated periodically. This database involves 1430 drugs and 5868 ADRs, resulting in 139,756 unique drug–ADR pairs [42]. We developed a reference standard wherein we regarded all drug–side effect pairs in SIDER as positive controls. The selection of negative controls was modeled by pairing each drug that appeared in the set of positive controls with one event that appeared in SIDER. We further removed each of the pairs that also appeared in the set of positive controls. Note that negative controls lack scientific support in this reference standard and might actually be positive controls. Furthermore, we restricted the evaluation to drug–ADR pairs for which Twitter contained at least one post and FAERS contained at least one case report. The minimum number of case counts was to ensure numeric stability in the signal detection estimates. Since Twitter was processed using three different symptom lists, the reference standard may vary when taking these three lists into account. Based on the three abovementioned reference standards, we compared the performance of the proposed combination system against that of signal scores generated by a baseline combination system and each data source independently. Performance was measured using the AUC. To test whether the differences in AUCs based on the different combination systems and individual systems were statistically significant, we computed a two-sided p-value under the null hypothesis that there is no difference between the AUCs of the two systems. The tests were computed using a bootstrapping method [43].
Baselines
To evaluate the proposed method, we compared it with the method proposed by Harpaz et al [44] which is also an empirical Bayesian method that combines ADR signals from multiple sources, where ADR signal scores mined from each data source are modeled concomitantly using a Bayesian two-stage normal/normal model whose two hyper-parameters are estimated from the data. Unlike our method, which takes a heterogeneous view by weighting each source according to their reliability measured by the score variance within each data source, it considers different data sources homogeneously.
Results
We acquired four data sets for further signal analysis and synthesis: Twitter Wiki, Twitter UMLS1, Twitter UMLS2, and FAERS. The characteristics of these four data sets are reported in Table 2. Using the Wiki symptom list obtained fewer ADRs (e.g., 40) than using the UMLS symptom lists (55 and 69, respectively). Using different symptom lists affected the number of drugs, the number of tweets, and consequently the derived statistics such as number of unique drug–ADR pairs indirectly since we implemented a rule that a tweet should mention both a symptom (“disease” or “symptom”) and a drug (“pharmacological substance”). Specifically, the numbers of drugs increased from 286 to 298, the numbers of relevant tweets increased from 55,867 to 72,008, the numbers of drug–ADR pairs increased from 1626 to 2036 using Wiki, UMLS1, and UMLS2, respectively. Meanwhile, FAERS involved 2.3 million reports during the same time span covering 3639 drugs, 15,173 ADRs, and 2.4 million unique drug–ADR pairs. In general, FAERS had higher rates of drugs per ADR and ADRs per drug than did Twitter, indicating that FAERS has broader coverage regarding ADR reports. Twitter had a higher rate of drugs per ADR than of ADRs per drug, whereas FAERs had a higher rate of ADRs per drug than of drugs per ADR.
Table 2.
Summary statistics for four data sets
| Data source | Reports (N) | Drugs (N) | ADRs (N) | Drug–ADR pairs (N) | Drugs per ADRb | ADRs per drugb |
|---|---|---|---|---|---|---|
| Twitter Wiki | 55,867 | 286 | 40 | 1626 | 41.69 | 5.71 |
| Twitter UMLS1 | 64,195 | 290 | 55 | 1768 | 32.74 | 6.12 |
| Twitter UMLS2 | 72,008 | 298 | 69 | 2036 | 29.94 | 6.86 |
| FAERS | 2.3 milliona | 3639 | 15,173 | 2.4 milliona | 159.42 | 664.72 |
ADR adverse drug reaction, FAERS US FDA Adverse Event Reporting System, UMLS Unified Medical Language System
aNumber of drug–ADR pairs is bigger than the number of reports because multiple drugs and events were mentioned in a single case report
bDrugs per ADR is the average number of unique drugs that are mentioned with an ADR; ADRs per drug is the average number of unique ADRs that are mentioned with a drug
Table 3 shows the top ten most frequently reported drugs and ADRs. The top ten drugs were almost the same for the three Twitter data sources, with the only exception that pseudoephedrine was in the top ten for UMLS2 but aspirin was in top ten for the other two Twitter sources. The top ten ADRs varied more than the top ten drugs, as the three symptom lists used were directly applied to identify symptoms that were potential candidates for ADRs. The top ten drugs in FAERS differed from those in the Twitter sources, and only aspirin and acetaminophen appeared in both FAERS and Twitters. The top ten ADRs in FAERS overlapped with the Twitter data sources for four exact ADR terms (nausea, pain, headache, dizziness) and several similar terms (e.g., vomiting and emesis). Death is a serious ADR that only appeared in the top ten ADRs from FAERS.
Table 3.
The top ten most frequently reported drugs and adverse drug reactions in each data source
| Data Source | Twitter Wiki | Twitter UMLS1 | Twitter UMLS2 | FAERS |
|---|---|---|---|---|
| Top ten drugs | Acetaminophen | Acetaminophen | Acetaminophen | Aspirin |
| Hydrocodone | Hydrocodone | Hydrocodone | Etanercept | |
| Diphenhydramine | Diphenhydramine | Diphenhydramine | Adalimumab | |
| Oxycodone | Oxycodone | Oxycodone | levothyroxine | |
| Caffeine | Caffeine | Caffeine | Omeprazole | |
| Phenylephrine | Dextromethorphan | Dextromethorphan | Acetaminophen | |
| Dextromethorphan | Menthol | Phenylephrine | Amlodipine | |
| Menthol | Phenylephrine | Menthol | Furosemide | |
| Ibuprofen | Ibuprofen | Ibuprofen | Prednisone | |
| Aspirin | Aspirin | Pseudoephedrine | Multivitamin preparation | |
| Top ten ADRs | Pain | Pain | Pain | Nausea |
| Headache | Headache | Headache | Drug ineffective | |
| Dizziness | Dizziness | Dizziness | Fatigue | |
| Nausea | Nausea | Nausea | Dyspnea | |
| Sleepy | Itching | Drowsiness | Pain | |
| Itching | Emesis | Itching | Diarrhea | |
| Fainting | Fainting | Emesis | Headache | |
| Cough | Cough | Fainting | Death | |
| Back pain | Backache | Cough | Vomiting | |
| Back ache | Insomnia | Backache | Dizziness |
ADR adverse drug reaction, FAERS US FDA Adverse Event Reporting System, UMLS Unified Medical Language System
The ROC AUC evaluations were based on the drug–ADR pairs that occurred in Twitter, FAERS, and the reference standard. Thus, the numbers of positive controls and negative controls varied when we intersected one of the Twitter data sets with FAERS and the reference standard, as shown in Table 4. The signal scores generated based on FAERS alone and Twitter alone were measured using the lower 5th percentile of MCEM output. When evaluated against their related reference standards, the FAERS data alone always achieved the highest AUCs (0.642, 0.613, and 0.612) compared with Twitter Wiki, Twitter UMLS1, and Twitter UMLS2, respectively. The proposed combination resulted in AUCs of 0.637, 0.578, and 0.595, respectively. These numbers were higher than the AUCs of the baseline combination method across the board. The Twitter sources alone always had the worst AUCs (0.534, 0.532, and 0.525, respectively). The differences in AUCs for the three Twitter sources alone were small, although the evaluations were based on different reference standards.
Table 4.
The AUCs of signal detection performance for Twitter, FAERS, and combined systems using relevant reference standards
| Data source | Method | AUC | Positive controls (N) | Negative controls (N) |
|---|---|---|---|---|
| Twitter Wiki and FAERS | FAERS alone | 0.642 | 489 | 348 |
| Twitter alone | 0.534 | 489 | 348 | |
| Baseline combination | 0.603 | 489 | 348 | |
| Proposed combination | 0.637 | 489 | 348 | |
| Twitter UMLS1 and FAERS | FAERS alone | 0.613 | 455 | 390 |
| Twitter alone | 0.532 | 455 | 390 | |
| Baseline combination | 0.578 | 455 | 390 | |
| Proposed combination | 0.587 | 455 | 390 | |
| Twitter UMLS2 and FAERS | FAERS alone | 0.612 | 465 | 456 |
| Twitter alone | 0.525 | 465 | 456 | |
| Baseline combination | 0.572 | 465 | 456 | |
| Proposed combination | 0.595 | 465 | 456 |
ADR adverse drug reaction, AUC area under the receiver operating characteristics curve, FAERS US FDA Adverse Event Reporting System, UMLS Unified Medical Language System
The p-values in Table 5 indicate that AUC differences between FAERS data alone and Twitter data alone, and between FAERS data alone and the baseline combination, were statistically significant (e.g., their two-sided p values were < 0.05). The proposed combination system achieved a comparable AUC with FAERS alone when using Twitter Wiki and FAERS data sets but performed significantly worse in other scenarios. In general, the combination systems achieved better AUCs than Twitter, although some were not significant. Similarly, the proposed combination system achieved a significantly better AUC than the baseline method when using Twitter Wiki and FAERS data sets (p = 0.0013). Figure 2 shows the resulting ROC curves for the signal detection based on each individual data source and two combination systems.
Table 5.
Two-sided p values for the hypothesis test of no difference in AUC performance between two methods
| Data source | Method | Twitter alone | Baseline combination | Proposed combination |
|---|---|---|---|---|
| Twitter Wiki and FAERS | FAERS alone | 0.0005 | 0.0003 | 0.2037 |
| Twitter alone | – | 0.0422 | 0.0011 | |
| Baseline combination | – | – | 0.0013 | |
| Twitter UMLS1 and FAERS | FAERS alone | 0.0103 | 0.0103 | 0.0031 |
| Twitter alone | – | 0.1830 | 0.1096 | |
| Baseline combination | – | – | 0.4314 | |
| Twitter UMLS2 and FAERS | FAERS alone | 0.0029 | 0.0024 | 0.0106 |
| Twitter alone | – | 0.1665 | 0.0328 | |
| Baseline combination | – | – | 0.0563 |
ADR adverse drug reaction, AUC area under the receiver operating characteristics curve, FAERS US FDA Adverse Event Reporting System, UMLS Unified Medical Language System
Fig. 2.
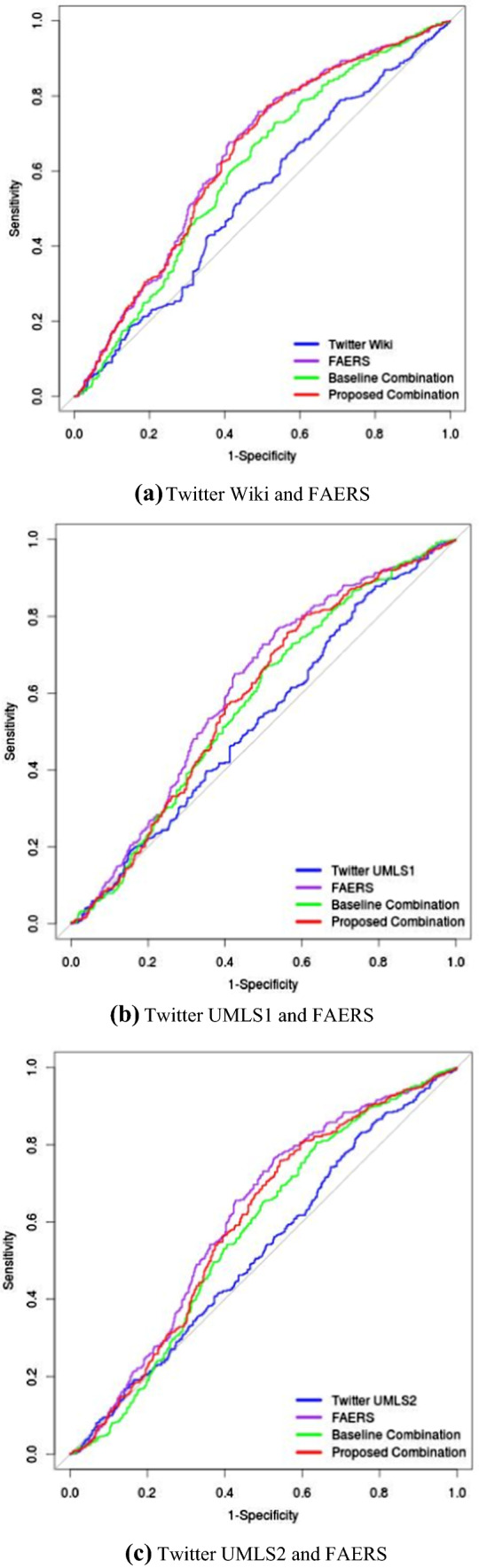
Receiver operating characteristic curves for signal scores based on Twitter, FAERS, and two combination systems. a Twitter Wiki, and FAERS; b Twitter UMLS1, and FAERS; c Twitter UMLS2, and FAERS. FAERS US FDA Adverse Event Reporting System, UMLS Unified Medical Language System
We further examined whether different data sources had an advantage when detecting a particular set of ADRs. Table 6 shows that different ADRs were more effectively detected by different systems. Note that the significance test was not conducted for these individual ADRs because of insufficient samples.
Table 6.
Adverse drug reactions with the best area under the receiver operating characteristics curve in one of three systems or are undetermined
| FAERS | Combination System | Undetermined | |
|---|---|---|---|
| Abdominal pain, burning sensation, constipation, dizziness, dry skin, flushing, hunger, nausea, rash, toothache, tremor | Anxiety disorder, back pain, chest pain, cough, fatigue, hunger, lethargy, pain, seizure, thirst | Agitation, anxiety, dizziness, fatigue, headache, insomnia, myalgia, nausea, vertigo | Abdominal discomfort, alcoholism, alopecia, amnesia, arthralgia, blindness, chills, drooling, dry eye, dry mouth, ear pain, ear pruritus, eye pruritus, flatulence, hypersomnia, malaise, overweight, sinus headache, sneezing, snoring, somnolence, starvation, stress, throat irritation, wheezing |
FAERS US FDA Adverse Event Reporting System
Discussion
Although the top reported ADRs were similar between Twitter and FAERS (Table 3), our study suggests that Twitter postings of drug-related AEs tend to focus on much fewer AEs (about 80) than in FAERS (about 15,000) and less-serious AEs, such as dizziness, pain, and nausea, which affect quality of life rather than being clinically serious and significant AEs. This is also why we could not evaluate the overall study using two well-known reference standards, namely OMOP reference standard [39] and time-indexed reference standard of ADRs [41], both of which focus more on serious and clinically significant ADRs.
As Freifeld et al. [45] suggested, AE reports from social media sources should not be pooled with those from conventional postmarketing sources since the influx of non-serious AEs may dilute the serious AEs. Our combination method avoids this pooling procedure at the case report level and can synthesize the analysis at the signal level. Overall, our combination system can boost the performance of signal detection based on Twitter data alone by leveraging information from FAERS. In addition, our combination system can achieve comparable AUCs with the FAERS for some combination data sets, although signal detection based on FAERS alone achieves the best AUC performance across the board. We must also understand that social media provides information in real time, whereas the first mention of an AE in FAERS might take significant time, e.g., several years, which supports the use of social media as a complementary source of adverse events.
The proposed combination system cannot achieve better performance by synthesizing the signal scores from FAERS and Twitter when compared with each data source alone. This result differs from those in our previous study, whereas the combined system can achieve significantly better AUCs than can FAERS or observational healthcare data such as EHRs and medical claims data alone [28]. This observation is consistent with those from other combination methods. It may indicate that the poor quality of information extracted from Twitter means that data are not ready to be integrated into any combination systems that are merely using data-driven methods. A possible way to improve the proposed combination system is to incorporate the expert knowledge through Bayesian probability theory by giving different weights to evidence from independent sources of information.[46].
This study has several limitations. First, our study only used Twitter data, the character restrictions on which may prevent users from discussing complex AEs. Thus, we are uncertain as to whether our findings could be generalized to other social media data sources such as patient forums. Second, the set of symptoms that our system identified was limited to self-reported symptoms that do not include ADRs identified in laboratory tests (e.g., blood test-derived ADRs). Third, the annotation method for processing Twitter data could not detect negated NE recognition. For example, a post that mentioned “I’m just not sleepy tonight” was annotated as “sleepy” (a potential AE symptom) by the NE tagger; however, the correct AE should be “insomnia”. This finding suggested that we needed to incorporate modification such as negation in the annotation method. Although the observation period for the Twitter data was from 2012 to 2014, reflecting a relatively aged data set, the overall combination system aimed to demonstrate the feasibility of using a statistical method to synthesize signals from FAERS and Twitter. This combination system could be generalized to combine FAERS with more recent Twitter data. Fourth, the current study design could not confirm whether Twitter could identify some ADR signals earlier than could traditional pharmacovigilance approaches. This requires a benchmark that can support prospective performance evaluations.
Conclusions
We presented a large-scale, efficient, and effective approach to systematically combine signals from Twitter and FAERS. Compared with signal detection solely using Twitter data, our combination system synthesizing signals from both FAERS and Twitter had significantly improved performance. However, given the several limitations associated with the data and reference standard used in this study, we cannot reach definitive conclusions regarding the usefulness of social media data to supplement conventional postmarketing surveillance. Future research directions involve incorporation of patient and health websites, expanding the scope of the reference standard, considering the time dimension of signal detection, and weighting evidence according to its fidelity.
Compliance with Ethical Standards
Funding
No sources of funding were used to conduct this study or prepare this manuscript. This project was supported by IBM Research.
Conflict of interest
Ying Li and Antonio Jimeno Yepes are employed by IBM Research. Cao Xiao is employed by IQVIA. All authors have no conflicts of interest that are directly relevant to the content of this study.
Data Sharing
The source data from FAERS for this study were extracted from another study that made its data publicly available (https://dx.doi.org/10.5061/dryad.8q0s4). The source data from Twitter cannot be shared because it was acquired under a commercial contract. The data set that we used to build the models for NLP is publicly available (https://github.com/IBMMRL/medinfo2015).
Footnotes
References
- 1.Hauben M, Madigan D, Gerrits CM, Walsh L, Van Puijenbroek EP. The role of data mining in pharmacovigilance. 2005. [DOI] [PubMed]
- 2.Powell GE, Seifert HA, Reblin T, Burstein PJ, Blowers J, Menius JA, et al. Social media listening for routine post-marketing safety surveillance. Drug Saf. 2016;39(5):443–454. doi: 10.1007/s40264-015-0385-6. [DOI] [PubMed] [Google Scholar]
- 3.van der Heijden PG, van Puijenbroek EP, van Buuren S, van der Hofstede JW. On the assessment of adverse drug reactions from spontaneous reporting systems: the influence of under-reporting on odds ratios. Stat Med. 2002;21(14):2027–2044. doi: 10.1002/sim.1157. [DOI] [PubMed] [Google Scholar]
- 4.Poluzzi E, Raschi E, Piccinni C, De Ponti F. Data mining techniques in pharmacovigilance: analysis of the publicly accessible FDA adverse event reporting system (AERS). Data mining applications in engineering and medicine. IntechOpen; 2012.
- 5.Ventola CL. Social media and health care professionals: benefits, risks, and best practices. Pharm Ther. 2014;39(7):491. [PMC free article] [PubMed] [Google Scholar]
- 6.Sarker A, Ginn R, Nikfarjam A, O’Connor K, Smith K, Jayaraman S, et al. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform. 2015;54:202–212. doi: 10.1016/j.jbi.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox S, Duggan M. Health online 2013. Health. 2013;2013:1–55. [Google Scholar]
- 8.Wittich CM, Burkle CM, Lanier WL, editors. Ten common questions (and their answers) about off-label drug use. Mayo Clinic Proceedings. Elsevier; 2012. [DOI] [PMC free article] [PubMed]
- 9.Kuehn BM. Scientists mine web search data to identify epidemics and adverse events. JAMA. 2013;309(18):1883–1884. doi: 10.1001/jama.2013.4015. [DOI] [PubMed] [Google Scholar]
- 10.Naik P, Umrath T, van Stekelenborg J, Ruben R, Abdul-Karim N, Boland R, et al. Regulatory definitions and good pharmacovigilance practices in social media: challenges and recommendations. Ther Innov Regul Sci. 2015;49(6):840–851. doi: 10.1177/2168479015587362. [DOI] [PubMed] [Google Scholar]
- 11.Food, Administration D. Guidance for industry: fulfilling regulatory requirements for postmarketing submissions of interactive promotional media for prescription human and animal drugs and biologics. 2014.
- 12.White RW, Harpaz R, Shah NH, DuMouchel W, Horvitz E. Toward enhanced pharmacovigilance using patient-generated data on the internet. Clin Pharmacol Ther. 2014;96(2):239–246. doi: 10.1038/clpt.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norén GN. Pharmacovigilance for a revolving world: prospects of patient-generated data on the internet. Springer; 2014. [DOI] [PubMed]
- 14.MacKinlay A, Aamer H, Yepes AJ, editors. Detection of adverse drug reactions using medical named entities on Twitter. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2017. [PMC free article] [PubMed]
- 15.Frost J, Okun S, Vaughan T, Heywood J, Wicks P. Patient-reported outcomes as a source of evidence in off-label prescribing: analysis of data from PatientsLikeMe. J Med Internet Res. 2011;13(1):e6. doi: 10.2196/jmir.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Chen H. A research framework for pharmacovigilance in health social media: identification and evaluation of patient adverse drug event reports. J Biomed Inform. 2015;58:268–279. doi: 10.1016/j.jbi.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Schröder S, Zöllner YF, Schaefer M. Drug related problems with antiparkinsonian agents: consumer internet reports versus published data. Pharmacoepidemiol Drug Saf. 2007;16(10):1161–1166. doi: 10.1002/pds.1415. [DOI] [PubMed] [Google Scholar]
- 19.Pages A, Bondon-Guitton E, Montastruc JL, Bagheri H. Undesirable effects related to oral antineoplastic drugs: comparison between patients’ internet narratives and a national pharmacovigilance database. Drug Saf. 2014;37(8):629–637. doi: 10.1007/s40264-014-0203-6. [DOI] [PubMed] [Google Scholar]
- 20.Pierce CE, Bouri K, Pamer C, Proestel S, Rodriguez HW, Van Le H, et al. Evaluation of Facebook and Twitter monitoring to detect safety signals for medical products: an analysis of recent FDA safety alerts. Drug Saf. 2017;40(4):317–331. doi: 10.1007/s40264-016-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duh MS, Cremieux P, Audenrode MV, Vekeman F, Karner P, Zhang H, et al. Can social media data lead to earlier detection of drug-related adverse events? Pharmacoepidemiol Drug Saf. 2016;25(12):1425–1433. doi: 10.1002/pds.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Ryan PB, Wei Y, Friedman C. A method to combine signals from spontaneous reporting systems and observational healthcare data to detect adverse drug reactions. Drug Saf. 2015;38(10):895–908. doi: 10.1007/s40264-015-0314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harpaz R, DuMouchel W, Schuemie M, Bodenreider O, Friedman C, Horvitz E, et al. Toward multimodal signal detection of adverse drug reactions. J Biomed Inform. 2017;76:41–49. doi: 10.1016/j.jbi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harpaz R, Vilar S, DuMouchel W, Salmasian H, Haerian K, Shah NH, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc. 2013;20(3):413–419. doi: 10.1136/amiajnl-2012-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, Wang Q. Large-scale combining signals from both biomedical literature and the FDA Adverse Event Reporting System (FAERS) to improve post-marketing drug safety signal detection. BMC Bioinform. 2014;15(1):17. doi: 10.1186/1471-2105-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilar S, Harpaz R, Chase HS, Costanzi S, Rabadan R, Friedman C. Facilitating adverse drug event detection in pharmacovigilance databases using molecular structure similarity: application to rhabdomyolysis. J Am Med Inform Assoc. 2011;18(Suppl 1):i73–i80. doi: 10.1136/amiajnl-2011-000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccinni C, Poluzzi E, Orsini M, Bergamaschi S, editors. PV-OWL—Pharmacovigilance surveillance through semantic web-based platform for continuous and integrated monitoring of drug-related adverse effects in open data sources and social media. 2017 IEEE 3rd International Forum on Research and Technologies for Society and Industry (RTSI). IEEE; 2017.
- 28.Xiao C, Li Y, Baytas IM, Zhou J, Wang F. An MCEM framework for drug safety signal detection and combination from heterogeneous real world evidence. Sci Rep. 2018;8(1):1806. doi: 10.1038/s41598-018-19979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banda JM, Evans L, Vanguri RS, Tatonetti NP, Ryan PB, Shah NH. A curated and standardized adverse drug event resource to accelerate drug safety research. Sci Data. 2016;3:160026. doi: 10.1038/sdata.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng QT, Tse T. Exploring and developing consumer health vocabularies. J Am Med Inform Assoc. 2006;13(1):24–29. doi: 10.1197/jamia.M1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikipedia. List of medical symptoms. https://en.wikipedia.org/wiki/List_of_medical_symptoms
- 33.Yom-Tov E, Gabrilovich E. Postmarket drug surveillance without trial costs: discovery of adverse drug reactions through large-scale analysis of web search queries. J Med Internet Res. 2013;15(6):e124. doi: 10.2196/jmir.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jimeno-Yepes A, MacKinlay A, Han B, Chen Q. Identifying diseases, drugs, and symptoms in Twitter. 2015. [PubMed]
- 36.Lafferty J, McCallum A, Pereira FC. Conditional random fields: probabilistic models for segmenting and labeling sequence data. 2001.
- 37.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Pham M, Cheng F, Ramachandran K. A comparison study of algorithms to detect drug-adverse event associations: frequentist, Bayesian, and machine-learning approaches. Drug Saf. 2019;42(6):743–750. doi: 10.1007/s40264-018-00792-0. [DOI] [PubMed] [Google Scholar]
- 39.Ryan PB, Schuemie MJ, Welebob E, Duke J, Valentine S, Hartzema AG. Defining a reference set to support methodological research in drug safety. Drug Saf. 2013;36(1):33–47. doi: 10.1007/s40264-013-0097-8. [DOI] [PubMed] [Google Scholar]
- 40.Coloma PM, Avillach P, Salvo F, Schuemie MJ, Ferrajolo C, Pariente A, et al. A reference standard for evaluation of methods for drug safety signal detection using electronic healthcare record databases. Drug Saf. 2013;36(1):13–23. doi: 10.1007/s40264-012-0002-x. [DOI] [PubMed] [Google Scholar]
- 41.Harpaz R, Odgers D, Gaskin G, DuMouchel W, Winnenburg R, Bodenreider O, et al. A time-indexed reference standard of adverse drug reactions. Sci Data. 2014;1. [DOI] [PMC free article] [PubMed]
- 42.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2015;44(D1):D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harpaz R, DuMouchel W, LePendu P, Shah NH. Empirical Bayes model to combine signals of adverse drug reactions. In: Proceedings of the 19th ACM SIGKDD International Conference on Knowledge discovery and data mining 2013 Aug 11, pp. 1339–1347
- 45.Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T, et al. Digital drug safety surveillance: monitoring pharmaceutical products in Twitter. Drug Saf. 2014;37(5):343–350. doi: 10.1007/s40264-014-0155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao C, Li Y, Argentinis E, Zhou J, Wang F, editors. An MCEM-MTL Framework for Drug Safety Signal Filtering and Detection in Spontaneous Reporting Systems [abstract ISoP18-1153]. In: 18th ISoP annual meeting “pharmacovigilance without borders” Geneva, Switzerland, 11–14 November, 2018. Drug Saf 2018;41:1103–1273