SUMMARY
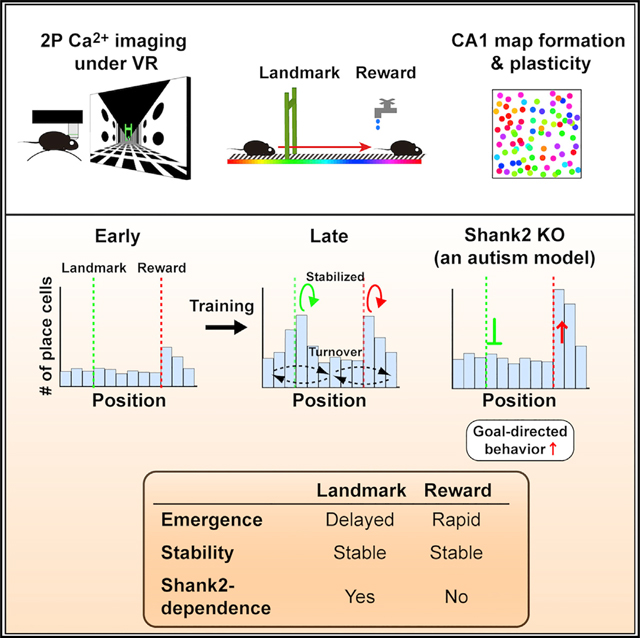
In the hippocampus, locations associated with salient features are represented by a disproportionately large number of neurons, but the cellular and molecular mechanisms underlying this over-representation remain elusive. Using longitudinal calcium imaging in mice learning to navigate in virtual reality, we find that the over-representation of reward and landmark locations are mediated by persistent and separable subsets of neurons, with distinct time courses of emergence and differing underlying molecular mechanisms. Strikingly, we find that in mice lacking Shank2, an autism spectrum disorder (ASD)-linked gene encoding an excitatory postsynaptic scaffold protein, the learning-induced over-representation of landmarks was absent whereas the over-representation of rewards was substantially increased, as was goal-directed behavior. These findings demonstrate that multiple hippocampal coding processes for unique types of salient features are distinguished by a Shank2-dependent mechanism and suggest that abnormally distorted hippocampal salience mapping may underlie cognitive and behavioral abnormalities in a subset of ASDs.
In Brief
Using longitudinal two-photon calcium imaging in mice during virtual navigation, Sato et al. demonstrate that persistent and separable neuronal subsets mediate the hippocampal over-representation of reward and landmark locations. Learning-induced over-representation of landmarks is absent while rapid over-representation of rewards is enhanced, in a mouse model of autism lacking Shank2.
Graphical abstract
INTRODUCTION
Navigation and spatial memory are essential elements of animal behavior that allow animals to forage, return home, and avoid danger. The hippocampus plays a crucial role in these cognitive processes, as hippocampal neurons fire when an animal is located in a particular part of an environment, providing an allocentric cognitive map of space (O’Keefe and Nadel, 1978). Although whether these “place cells” (PCs) are indeed memory cells has been long debated, one line of evidence that favors this notion indicates that hippocampal place-specific firing exhibits dynamic changes according to context and experience on multiple timescales, ranging from a few minutes to days or weeks (Muller and Kubie, 1987; Bostock et al., 1991; Mehta et al., 1997; Lever et al., 2002; Leutgeb et al., 2005). Furthermore, studies have reported that disproportionately large numbers of PCs are recorded in locations that are associated with reward, safety, or local cues (O’Keefe and Conway, 1978; Wiener et al., 1989; Hetherington and Shapiro, 1997; Hollup et al., 2001; Dupret et al., 2010; Danielson et al., 2016; Zaremba et al., 2017; Gauthier and Tank, 2018, Bourboulou et al., 2019), indicating that the environment surrounding an animal is not represented uniformly in the hippocampal cognitive map; representations are strongly influenced by the motivational and environmental salience of the locations.
These findings imply that the activity of an increased number of neurons encodes the presence of salience in the hippocampal map. This idea further proposes potential roles of such salience maps not only in spatial (Hollup et al., 2001; Dupret et al., 2010) or episodic-like memories (Komorowski et al., 2009; Eichenbaum and Cohen, 2014) but also in goal-directed and landmark-based navigation (Burgess and O’Keefe, 1996; Gothard et al., 1996). PCs are formed rapidly within minutes after initial exposure to a new environment (Hill, 1978; Wilson and McNaughton, 1993; Frank et al., 2004). However, how the over-representation of salient features is established and modified by experience remains to be fully understood. Several important questions regarding these salience maps remain to be addressed: for example, whether the over-representation of reward and that of other types of salient features are mediated by a single shared or multiple distinct mechanisms and how altered over-representation relates to behavioral and cognitive abnormalities in various brain disorders, including autism spectrum disorders (ASDs).
To elucidate the cellular and molecular mechanisms that govern the dynamics of hippocampal salience representation, we longitudinally imaged functional cellular maps of the deep sublayer of CA1 during training on a virtual linear track, in which two distinct locations were associated with reward or a visual landmark. We show that over-representations of motivationally and environmentally salient features are mediated by persistent and separable subsets of neurons with distinct time courses of emergence and differing molecular mechanisms. Remarkably, mice lacking Shank2 (Won et al., 2012), a mouse model of ASD that lacks a glutamatergic postsynaptic scaffold protein, exhibit selective loss of learning-induced over-representation of landmark locations, while their rapid over-representation of reward locations and goal-directed behavior is further enhanced.
RESULTS
Mice and Behavioral Task
To reliably perform longitudinal imaging of large-scale functional hippocampal cellular maps, we generated a transgenic mouse line, herein termed Thy1-G-CaMP7, that coexpresses the fluorescent calcium indicator protein G-CaMP7 and the calcium-insensitive red fluorescent marker protein DsRed2 via 2A peptide-mediated bicistronic expression under the neuron-specific Thy1 promoter (Figure 1A; Ohkura et al., 2012; Sato et al., 2015; see also STAR Methods and Figure S1). In the dorsal CA1 of the hippocampus, the population of calbindin D-28K-negative pyramidal cells in the deep pyramidal cell sublayer was preferentially labeled with G-CaMP7 (Mizuseki et al., 2011; Kohara et al., 2014; Lee et al., 2014; Valero et al., 2015; Danielson et al., 2016; Figures 1B and S1A). Immunofluorescence labeling of glutamic acid decarboxylase 65/67, parvalbumin, and somatostatin revealed that interneurons positive for these markers were devoid of G-CaMP7 expression (Figure S1B).
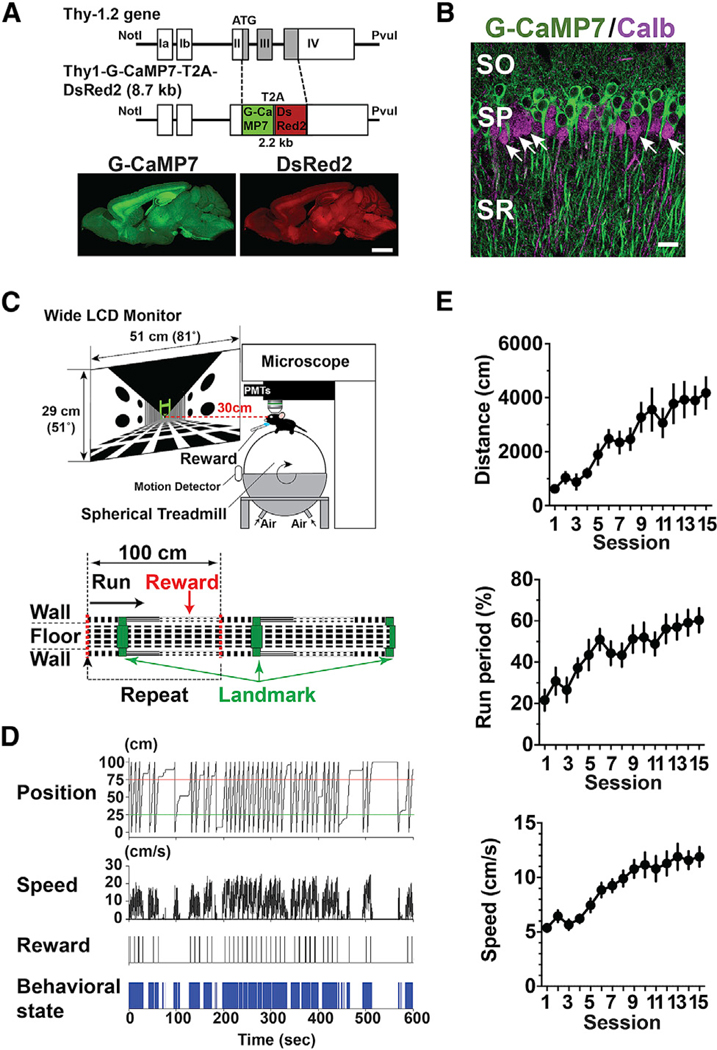
Figure 1. Transgenic Mice and Behavioral Task.
(A) Transgene construct for Thy1-G-CaMP7 mice (top) and expression of G-CaMP7 (bottom left, green) and DsRed2 (bottom right, red) in a parasagittal section from a mouse at six months of age. Scale bar, 2 mm.
(B) G-CaMP7 expression (green) and calbindin immunofluorescence (calb, magenta) in the dorsal CA1 of the hippocampus of Thy1-G-CaMP7 transgenic mice. Arrows indicate examples of calbindin-positive G-CaMP7-negative cells. SO, stratum oriens; SP, stratum pyramidale; SR, stratum radiatum. Scale bar, 20 μm.
(C) A schematic of the two-photon microscope and virtual reality setup (top) and virtual endless linear track task (bottom). The linear track segment contained a visual landmark (a green gate) and a reward delivery point at two distinct locations. When the mouse’s virtual position reached the point indicated by the red dotted line in the middle, it returned to the origin, such that the same track segment was presented repeatedly.
(D) Example behavioral data from a single 10-min session. From top to bottom, the mouse’s virtual position on the linear track, running speed, timing of reward delivery, and behavioral state are shown, where running is represented in blue.
(E) Behavioral changes induced by repeated training. Total distance traveled (distance, top), the fraction of time spent running (run period, middle), and running speed (speed, bottom) are shown. Data are expressed as mean ± SEM.
To allow imaging of hippocampal maps during repeated training of spatial behavior, we trained Thy1-G-CaMP7 mice in a virtual linear track task (Figures 1C and 1D; see STAR Methods for details). The mouse started running from the origin of the segment, passed under a green gate as a visual landmark, then received water at a reward point and returned to the origin instantaneously after reaching the other end. The visual landmark and reward delivery were associated with two distinct locations to examine the effects of two different types of salience separately. In training, behavioral performance as measured by time spent running, distance traveled, and running speed during 10-min sessions markedly increased as training proceeded (Figure 1E; distance, p < 0.0001, F(14,154) = 7.30; run period, p < 0.0001, F(14,154) = 5.26; speed, p < 0.0001, F(14,154) = 11.6; n = 12 mice from three groups, one-way analysis of variance [ANOVA]). Furthermore, licking and slowing of running speed before the reward delivery point developed as training proceeded (Figure S2). The emergence of such anticipatory behavior indicates that the task involves goal-directed spatial learning.
Rapid and Delayed Emergence Distinguishes Over-Representations of the Reward and Landmark Locations
To examine whether and when the representations of the two salient locations become prominent in the map, we next visualized the emergence and establishment of hippocampal CA1 spatial maps during training on the virtual linear track task (Figure S3; see STAR Methods for details on imaging and analysis). The cells that exhibited virtual-location-specific activity (PCs) showed spatially more informative activity and higher event frequency than non-PCs (Figures S3H–S3K).
Consistent with previous studies in real and virtual environments, PCs were formed rapidly within the first session on the virtual linear track (Hill, 1978; Wilson and McNaughton, 1993; Frank et al., 2004; Chen et al., 2013) (Figures 2A and 2B). The fractions of PCs were initially low but then increased as the training proceeded. The fraction of PCs and that of time spent running showed a good overall correlation (Figure 2C). The slope of a regression line (termed the “PC formation factor”) significantly increased in the late phase of training compared with the early phase (Figures S4A and S4B). Furthermore, the sessions in the late phase contained larger fractions of PCs than the sessions in the early phase with comparable amounts of running time (Figures S4C and S4D). The average event frequency, mutual information content, and response amplitude of PCs did not notably change during training (Figures S4E–S4G).
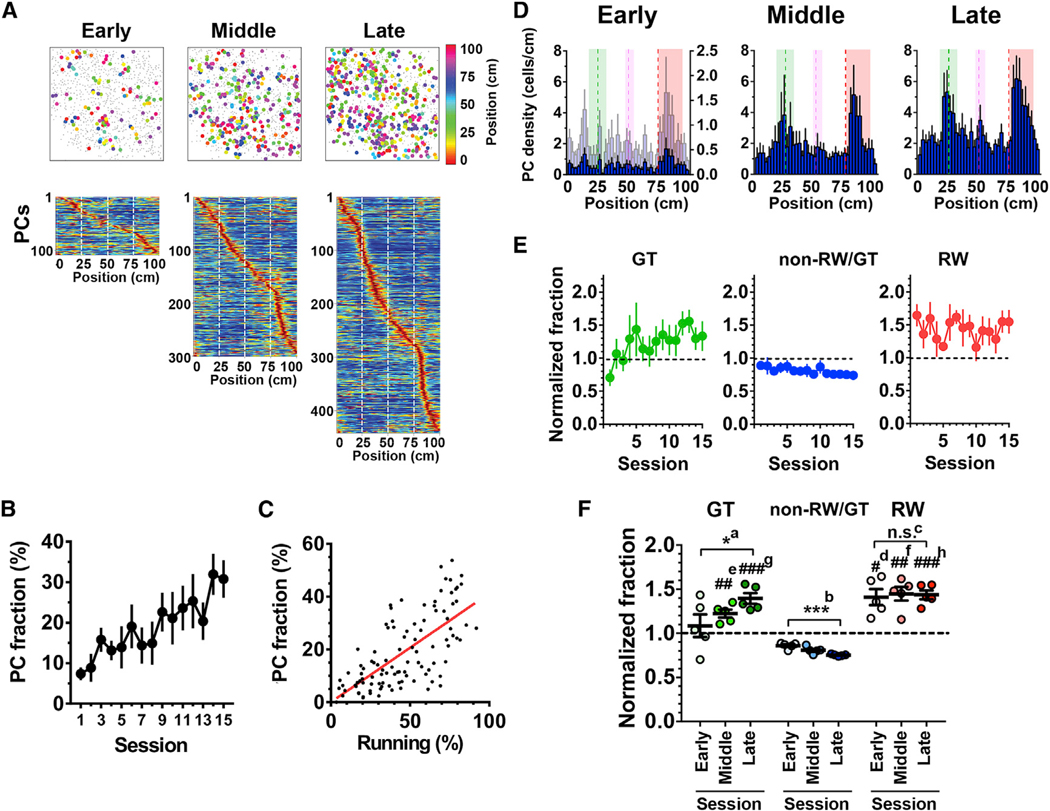
Figure 2. Over-Representations of the Reward and Landmark Locations Emerge with Different Time Courses.
(A) Examples of place cell (PC) maps imaged in the same animal at the early (session 1), middle (session 9), and late (session 14) phases of training on the virtual endless linear track task (top). PCs and non-PCs are represented by filled circles of various colors and gray dots, respectively. The different colors of the filled circles represent different locations of the place fields. Heatmaps shown below are distributions of place fields of the corresponding sessions ordered by their positions (bottom).
(B) The fractions of PCs relative to the number of total identified cells imaged at each session. n = 7 mice.
(C) The fractions of PCs were plotted against the corresponding fractions of time spent running (n = 105 sessions from seven mice). The red line represents linear regression (r = 0.59).
(D) Histograms indicating the distribution of PCs with respect to track position in the early (session 1), middle (session 6), and late (session 12) phases of the training. The average data from seven mice are shown. For comparison, the histogram of the early phase was scaled to that of the late phase by their maximum values and is plotted in light blue on its right y axis. The green, red, and magenta dashed lines delineate the positions of the landmark, reward delivery, and boundary between different wall patterns, respectively. The areas shown in green, red, and magenta indicate those that define gate, reward, and wall cells, respectively.
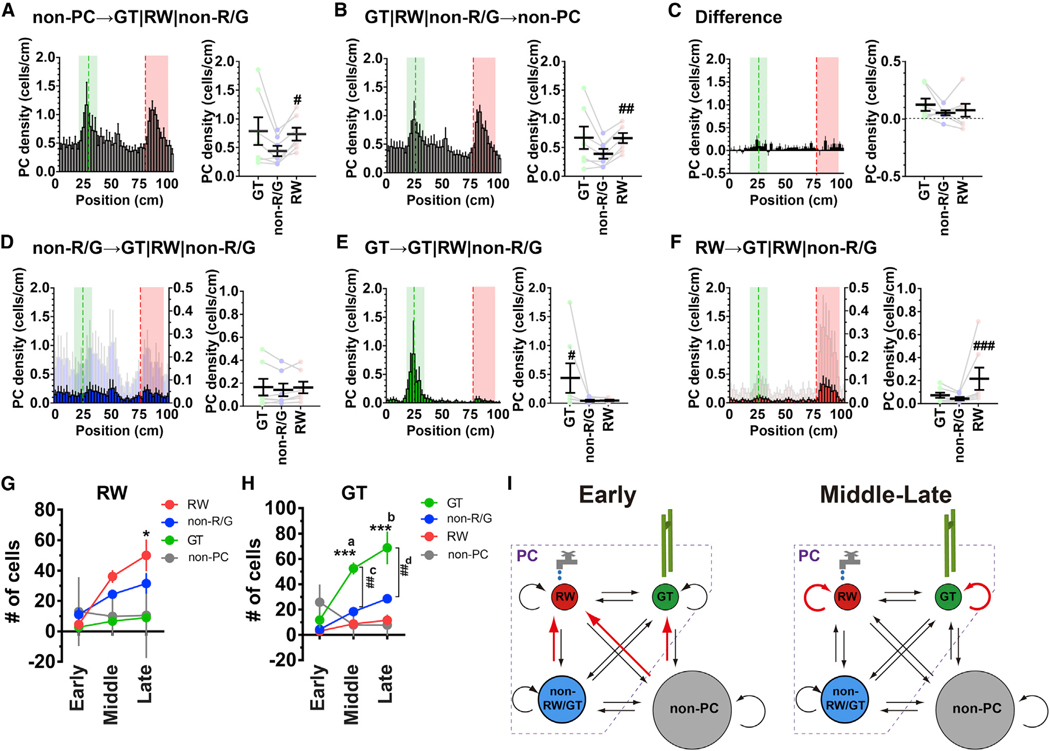
(E) Hippocampal spatial representations as expressed by the fractions of gate cells (GT cells, green), non-reward/gate cells (non-RW/GT cells, blue), and reward cells (RW cells, red) relative to the number of total PCs identified in each session. Values were normalized to that obtained in the case of uniform distribution (i.e., 0.0125/bin), and values greater than 1 indicate that the locations are over-represented.
(F) Average normalized fractions of GT (green), non-RW/GT (blue), and RW (red) cells for the early, middle, and late phases of the training. *a, p = 0.046, F(2,12) = 3.26; ***b, p = 0.0003, F(2,12) = 15.3; n.s.c, p = 0.96, F(2,12) = 0.067; one-way ANOVA, n = 5 sessions each; #d, p = 0.014 versus non-RW/GT early, F(1.059,4.235) = 6.33; ##e, p = 0.0037 versus non-RW/GT middle; ##f, p = 0.0044 versus non-RW/GT middle, F(1.334,5.335) = 27.8; ###g, p = 0.0009 versus non-RW/GT late; ###h, p= 0.0007 versus non-RW/GT late, F(1.142,4.566) = 50.68; one-way ANOVA; n = 5 sessions each.
(B and D–F) Data are expressed as mean ± SEM.
We then examined whether the locations associated with salience were disproportionately represented in the hippocampal map. The histograms of PCs against positions typically exhibited two large peaks, which appeared more clearly in the late training phase; one peak corresponded to the location of the landmark, and the other corresponded to that of the reward (Figure 2D). While the first peak closely matched the landmark location, the second peak was slightly shifted in the direction the mouse was running, which likely reflects that the animals received the rewards in places that were slightly past the delivery point, as suggested by decreased running speed in this area (Figure S2A). Importantly, the over-representation of the reward location was discernible even in the first session of training, whereas that of the landmark location gradually developed as the training proceeded (Figures 2D–2F). The fraction of PCs that encoded the location of the reward (herein termed “reward cells” [RW cells] for convenience) was not significantly different between the early and late phases of training, whereas that of PCs that encoded the location of the landmark (similarly termed “gate cells” [GT cells]) increased significantly, with a complementary decrease in the fraction of PCs that encoded other locations (termed “non-reward/gate cells” [non-RW/GT cells]; Figures 2E and 2F). The delayed emergence of increase in PCs that encode locations associated with salient visual cues is further supported by a more delayed and reduced increase in PCs that encode a location with less visual salience, such as a boundary of different wall patterns (termed “wall cells” [WL cells]; Figures S4H and S4I). Collectively, these results demonstrate that the over-representation of salient locations is formed and maintained at a population level, although the maps develop dynamically throughout the training period. The establishment and refinement of representations of salience depend on its nature; the representation of motivational salience is established rapidly, whereas that of environmental salience develops over the course of training.
RW Cells and GT Cells Form Stable Singularities during Experience-Dependent Map Consolidation
The PC maps imaged in each session appeared rather different from each other, even within the same animals, implying that hippocampal spatial representations are highly dynamic while being established (Figure 2A). To investigate whether representations of salient locations are more stable than those of nonsalient locations, we investigated training-induced changes in the maps at an individual cell level by comparing the place fields of the same cells across different sessions (Figure S5; see STAR Methods for details). In the early maps, only a small number of PCs were identified as common to both sessions (these cells are hereinafter called “common PCs”), but the fraction of common PCs increased significantly as additional PCs were imaged in the late phase of the training (Figures 3A–3C). Moreover, the fraction of PCs that had stable place fields in both sessions (“stable PCs”) also increased markedly as the training proceeded (Figures 3A, 3B, 3D, and 3E), indicating that the maps are consolidated in an experience-dependent manner. Image comparisons between adjacent sessions showed that the fractions of common cells were constant over time (p = 0.57, one-way ANOVA; Figure S5E). Furthermore, the fractions of common and stable PCs normalized to the number of PCs also increased significantly as the training proceeded, indicating that the training-induced increase in PC stability was not simply due to the increase in the number of PCs (Figure S6). We then calculated the fractions of stable PCs with respect to the number of common PCs as an index of stable representations at each location and found that this index was significantly higher for locations associated with the landmark or reward than for other locations (Figures 3F and 3G). In addition, the gain of PC stability between the early and late phases of training in individual animals exhibited a good linear correlation with their differences in time spent running between the two phases of the training (r = 0.90, Figure 3H). The results suggest that hippocampal place maps are more strongly stabilized if the animals learn to run the virtual linear track task more effectively.
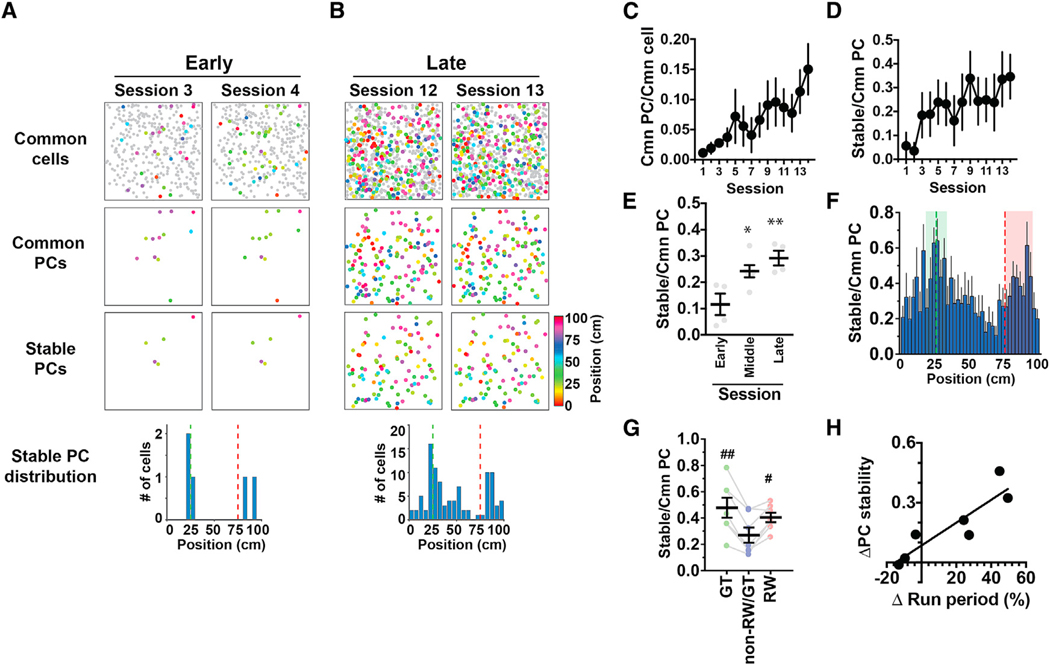
Figure 3. RW Cells and GT Cells Form Stable Singularities in Hippocampal Cognitive Maps.
(A) Example hippocampal CA1 PC maps imaged in two consecutive sessions in the early phase of the training. Maps shown on top, middle, and bottom present cells identified in common to both sessions (common cells), cells identified as PCs in both sessions (common PCs), and cells identified as PCs with stable (< 10 cm difference) place fields in both sessions (stable PCs), respectively. The histogram shown at the bottom indicates the distributions of the stable PCs against track position.
(B) PC maps imaged in the late phase of training in the same animal as presented in (A).
(C) The fraction of common PCs relative to the number of common cells identified in the two consecutive sessions that were compared. The x axis indicates the earlier of the two sessions that were compared. n = 7 mice.
(D) PC stability calculated as the fraction of stable PCs relative to the number of common PCs identified in the two consecutive sessions that were compared.
(E) Average PC stability in the early (sessions 1–4, numbered according to the earlier of the two sessions that were compared), middle (sessions 5–10), and late (sessions 11–14) phases of training. *p = 0.021 versus early, **p = 0.0050 versus early, F(2,11) = 8.04, one-way ANOVA; n = 4, 6, and 4 session pairs for the early, middle, and late phases, respectively.
(F) The average fractions of stable PCs relative to the number of common PCs plotted against track position. Values were calculated from data across all sessions and averaged for seven mice. The fractions were determined using the number of common PCs obtained separately for each spatial bin in the earlier of the two sessions compared. The green and red dashed lines delineate the positions of the landmark and reward delivery, respectively. The areas shown in green and red are those that define GT cells and RW cells, respectively.
(G) The average PC stability for GT, non-RW/GT, and RW cells. #p = 0.015 versus non-RW/GT cells, ##p = 0.0025 versus non-RW/GT cells, F(1.215,7.290) = 11.6, one-way ANOVA, n = 7 mice.
(H) The relationship between PC stability and learning to run along the track. The x axis presents the task performance of each mouse measured by the difference in the fraction of time spent running between the early (average of sessions 1–5) and late (average of sessions 11–15) phases of the training. The y axis presents the difference in PC stability between the early and late phases of training.
(C–G) Data are expressed as mean ± SEM.
Response Properties of RW Cells and GT Cells
In order to further characterize RW cells and GT cells, mice trained in the normal task were subjected to a task with 50% reward delivery and a task with no gate (n = 3 mice). In the 50% reward task, reward was delivered randomly with 50% probability at the same delivery point, and the data were separately analyzed for rewarded trials and non-rewarded trials (Figure 4A). The fraction of time spent running increased slightly but not significantly in non-rewarded trials (rewarded trials 66.9% ± 4.2% versus non-rewarded trials, 74.9% ± 3.9%, p = 0.076, t(5) = 2.23, paired two-tailed t test, n = 6 sessions from three mice). Licking within the reward zone decreased in non-rewarded trials with marginal statistical significance, whereas licking immediately before and far outside the reward zone was not altered between rewarded trials and non-rewarded trials (Figure 4B). Consistent with the notion that the presence of reward is encoded by the increased density of RW cells, the representation of the reward zone substantially decreased in non-rewarded trials (Figure 4C; rewarded trials, 1.19 ± 0.12 versus non-rewarded trials, 0.50 ± 0.09-fold relative to a uniform distribution [see Figure 2E legend], p = 0.0031, t(5) = 5.35, paired two-tailed t test, n = 6 sessions from three mice). Thus, RW cells appear to be enriched in the reward location due to the presence of reward.
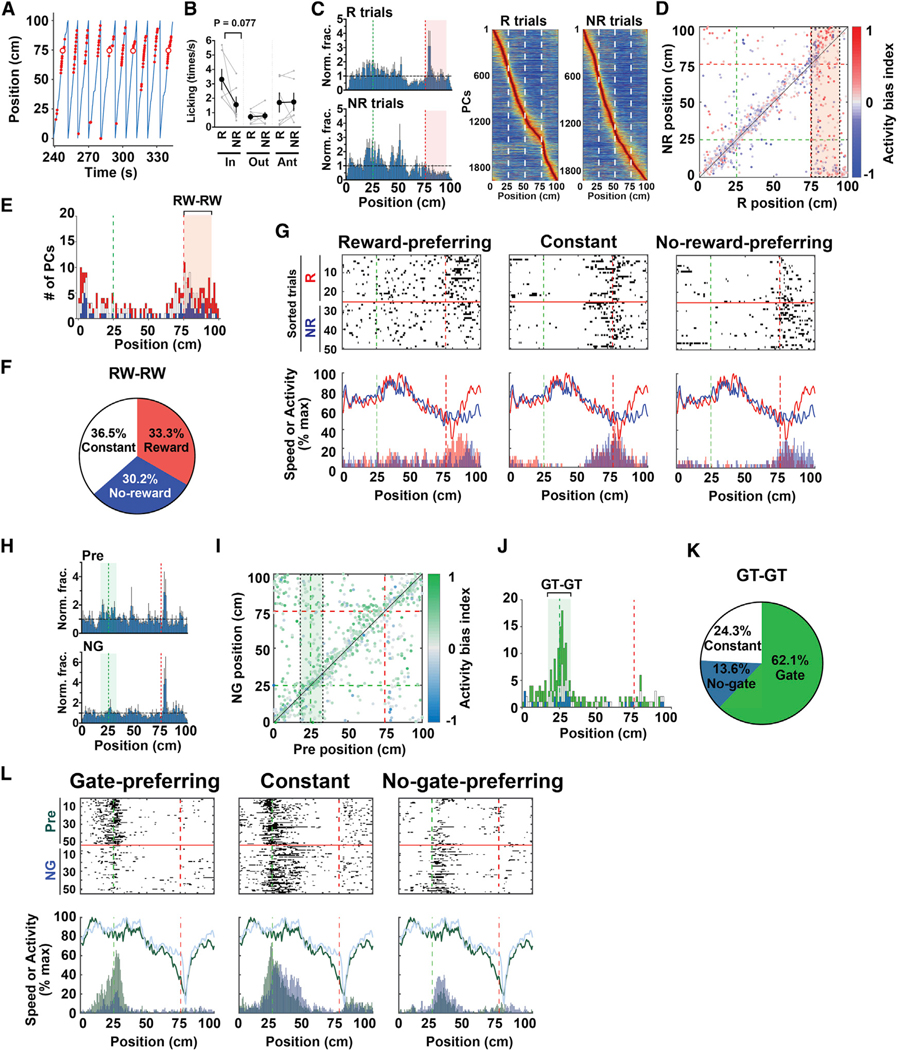
Figure 4. Response Properties of RW Cells and GT Cells.
(A) An example segment of 50% reward delivery sessions. The blue line represents the animal’s position. Red dots and red circles indicate licking and reward delivery, respectively.
(B) Licking within the reward zone (in), immediately before the reward zone (ant), and elsewhere (out) in rewarded (R) and non-rewarded (NR) trials (R in versus NR in, p = 0.077, t(5) = 2.22, paired two-tailed t test, n = 6 sessions each). Data are expressed as mean ± SEM.
(C) Histograms indicating the distribution of PCs with respect to track position for R and NR trials (left). The y axis represents PC fractions normalized relative to a uniform distribution. The average data of six sessions using three mice are shown. The green and red dashed lines delineate the positions of the landmark and reward delivery, respectively. The red area defines RW cells. Heatmaps shown on the right highlight the distributions of place fields in R and NR trials, ordered by their positions (right). n = 1,978 cells (R trials) and 1,915 cells (NR trials), respectively, from six sessions using three mice.
(D) A scatterplot showing the distribution of place-field positions in R and NR trials. Each dot represents the position of a cell’s place field, and the color indicates the activity bias index of the cell. Cells from all relevant sessions are shown. The green and red dashed lines delineate the positions of the landmark and reward delivery, respectively. The red area enclosed by a black dashed rectangle defines RW cells in R trials that remained PCs in NR trials, and a histogram of the distribution of their place fields along the NR axis is shown in (E). The same convention applies hereinafter.
(E) A histogram showing the distribution of place-field positions of PCs in NR trials that were RW cells in R trials. Red, white, and blue segments of the stacked bar represent the proportions of reward-preferring RW cells, constant RW cells, and no-reward-preferring RW cells, respectively, to the total number of cells in that bin. The red area defines RW cells in R trials that persisted as RW cells in NR trials (RW-RW), and the proportions of reward-preferring RW cells, constant RW cells, and no-reward-preferring RW cells in these cells are shown in (F).
(F) Proportions of reward-preferring RW cells, constant RW cells, and no-reward-preferring RW cells among RW-RW cells.
(G) Examples of reward-preferring RW cells (left), constant RW cells (middle), and no-reward-preferring RW cells (right). The three example cells shown were imaged during the same session. Top panels show raster plots against position, in which the plots for R and NR trials are divided by red horizontal lines and separately sorted. Bottom panels show activity histograms of each example cell against position for R (red) and NR (blue) trials and running speed averaged separately for R (red) and NR (blue) trials during the corresponding session.
(H) Histograms indicating the distribution of PCs with respect to track position for preceding control (pre) and no-gate (NG) sessions. The y axis represents PC fractions normalized relative to a uniform distribution. The average data from three sessions using three mice are shown. The green area defines GT cells.
(I) A scatterplot showing the distribution of place-field positions in pre and NG sessions. The green area enclosed by a black rectangle defines GT cells in pre sessions that remained PCs in NG sessions. A histogram of the distribution of their place fields along the NG axis is shown in (J).
(J) A histogram showing the distribution of place-field positions of PCs in NG sessions that were GT cells in pre sessions. Green, white, and turquoise segments of the stacked bar represent the proportions of gate-preferring GT cells, constant GT cells, and no-gate-preferring GT cells, respectively, to the total number of cells in that bin. The green area defines GT cells in pre that persisted as GT cells in the NG condition (GT-GT), and the proportions of gate-preferring GT cells, constant GT cells, and no-gate-preferring GT cells among these cells are shown in (K).
(K) Proportions of gate-preferring GT cells, constant GT cells, and no-gate-preferring GT cells among GT-GT cells.
(L) Examples of gate-preferring GT cells (left), constant GT cells (middle), and no-gate-preferring GT cells (right). Top panels show raster plots against position in which pre and NG sessions are shown above and below the red horizontal lines, respectively. Bottom panels show running speed averaged for pre (green) and NG (turquoise) sessions and activity histograms of each example cell against position for pre (green) and NG (turquoise) sessions.
We then tracked the changes in RW cells in non-rewarded trials. Twenty-one percent of RW cells (96 out of 467 cells) still had their place fields within the reward zone in non-rewarded trials (RW-RW cells; Figures 4D and 4E), while 37% (173 cells) became PCs that encoded other parts of the track and 42% (198 cells) lost their properties as PCs. The consistency of RW cells was markedly lower than the other categories of cells; 59% of GT cells (251 out of 423 cells) and 51% of non-RW/GT cells (552 out of 1,088 cells) in rewarded trials remained GT cells and non-RW/GT cells, respectively, in non-rewarded trials. However, 1.78-fold enrichment of RW–RW cells relative to a uniform distribution of PCs (96 out of 269 cells; Figure 4E) indicates that a part of RW cells still tend to be RW cells even in the absence of reward, which likely reflects a general tendency of PCs that their fields tend to remain in the same locations in non-rewarded trials (Figure 4D).
To further investigate how reward modulates RW cell activity, we compared the activity of each RW-RW cell between rewarded and non-rewarded trials and divided them into three groups according to the direction and extent of their activity biases (Figures 4F and 4G). Interestingly, we found that 30.2% of RW-RW cells (29 out of 96 cells) increased their activity in the absence of reward (no-reward-preferring RW cells), while approximately similar proportions of cells exhibited constant (termed “constant RW cells” after their constant activity regardless of the presence or absence of reward; this terminology does not mean they continuously encode the reward location, 36.5%) or decreased activity (reward-preferring RW cells, 33.3%; Figures 4F and 4G).
Next, we examined the effect of the visual landmark on GT cell activity in the no-gate task, in which the green gate was removed from its location throughout a test session that was conducted immediately after a preceding control session (pre). The fraction of time spent running did not change notably in no-gate sessions (pre, 76.4% ± 8.9%, versus no gate, 79.8% ± 9.5%, n = 3 sessions each from three mice). The representation of GT location decreased in no-gate sessions (Figure 4H; pre, 1.37 ± 0.19, versus no gate, 1.01 ± 0.06-fold relative to a uniform distribution, n = 3 sessions each), supporting the idea that the increased density of GT cells encodes the presence of the landmark. In no-gate sessions, 36% of GT cells identified in pre (103 out of 287 cells, 3.3-fold relative to a uniform distribution) maintained their place fields within the area that defined GT cells (GT-GT cells; Figures 4I and 4J), while 36% (104 out of 287 cells) had significant place fields in other parts of the track and the remaining 28% (80 out of 287 cells) lost their properties as PCs. Thus, similar to RW cells, GT cells appear to accumulate at the landmark location when the landmark is present. Moreover, a part of GT cells tend to be GT cells in the absence of landmark as PCs generally tend to have their fields at the same locations (Figures 4I and 4J). GT-GT cells were then categorized similarly into three groups by comparing their activity between the preceding control and no-gate sessions (Figures 4K and 4L). Notably, 62.1% (64 out of 103 cells) of GT-GT cells were gate-preferring GT cells, whereas only 13.6% of them (14 out of 103 cells) were no-gate-preferring GT cells (Figures 4J and 4K). This asymmetric modulation of GT cells by visual landmark indicates that the majority of GT cells decrease their activity when the visual landmark is not available.
Persistent Subsets of Cells Mediate Over-Representations of Reward and Landmark Locations
To elucidate the cellular mechanism of map establishment, we analyzed the transitions among RW, GT, non-RW/GT cells, and non-PCs between adjacent sessions (Figure 5). Although the conversion of PCs from non-PCs exhibited significant biases toward RW cells (Figure 5A), a subpopulation of PCs that became non-PCs also exhibited a similarly biased distribution (Figure 5B). This implies that the disproportionate formation of PCs is equilibrated by a similar bias in the disappearance of part of the PCs (Figure 5C). The distribution of PCs derived from former non-RW/GT cells appeared not to be significantly biased toward RW or GT cells (Figure 5D). In contrast, the distribution of PCs derived from former GT or RW cells was substantially biased toward the location by which each of the two PC subcategories was defined (Figures 5E and 5F). The persistence and relative independence of RW cells and GT cells demonstrate that selective stabilization of RW and GT cell subsets, rather than lateral recruitment of non-RW/GT cells, is a major mechanism that underlies the establishment of hippocampal salience maps. A further analysis of the cellular origins of RW cells and GT cells confirmed that most RW cells and GT cells in the middle and late sessions were cells of the same subcategories in preceding sessions, although these cells in the early sessions mainly originated from net formation from non-PCs (Figures 5G and 5H). Collectively, these findings indicate that hippocampal map formation consists of two steps: first, early populations of RW cells and GT cells are primarily derived from non-PCs until the number of cells is sufficient; second, thereafter, each subcategory becomes its own dominant source in the middle and late sessions (Figure 5I).
Figure 5. Persistent Neuronal Subsets Mediate Over-Representations of the Reward and Landmark Locations.
(A) Formation of different PC categories from non-PCs. (Left) A histogram showing the distribution of PCs that were non-PCs in the previous sessions against position. The values were calculated from data across all sessions and averaged for seven mice. For comparison, the histograms shown in (A), (B), and (D–F) are plotted on the left y axes on the same scale. In addition, the histograms in (D) and (F) were scaled by their maximum values and are plotted in a light color on the right y axes. (Right) The average cell density of each PC subcategory formed from non-PCs. #p = 0.023 versus non-RW/GT, χ2(2) = 8.00; Friedman test, n = 7 mice.
(B) Elimination of different PC categories by conversion to non-PCs. (Left) A histogram showing the distribution of PCs that became non-PCs in subsequent sessions against position. (Right) The average cell density of each PC subcategory that became non-PCs in the subsequent sessions. ##p = 0.0099 versus non-RW/GT, χ2(2) = 8.86; Friedman test, n = 7 mice.
(C) Net PC formation. (Left) A histogram of the difference obtained by subtracting the histogram in (B) from that in (A). (Right) The average cell density of PCs of each subcategory.
(D) Transition and stability of non-RW/GT cells. (Left) A histogram showing the distribution of PCs that were non-RW/GT cells in the previous sessions against position. (Right) The average cell density of each PC subcategory that was derived from non-RW/GT cells.
(E) Transition and stability of GT cells. (Left) A histogram showing the distribution of PCs that were GT cells in the previous sessions against position. (Right) The average cell density of each PC subcategory that was derived from GT cells. #p = 0.023 versus non-RW/GT, χ2(2) = 7.14, Friedman test, n = 7 mice.
(F) Transition and stability of RW cells. (Left) A histogram showing the distribution of PCs that were RW cells in the previous sessions against track position. (Right) The average cell density of each PC subcategory that was derived from RW cells. ###p = 0.0005 versus non-RW/GT, χ2(2) = 14.0, Friedman test, n = 7 mice.
(G) The numbers of RW cells that were derived from RW cells, GT cells, non-RW/GT cells, and non-PCs during the early, middle, and late phases of training. The transitions from non-PCs represent net formation after subtracting the disappearance of the existing PCs. *p = 0.021 versus early RW, F(3,33) = 3.16; two-way ANOVA, n = 4–6 session pairs.
(H) The numbers of GT cells that were derived from RW cells, GT cells, non-RW/GT cells, and non-PCs during the early, middle, and late phases of training. ***a, p = 0.0003 versus early GT, ***b, p < 0.0001 versus early GT, F(3,33) = 17.6; ##c, p = 0.0011 versus middle non-RW/GT, ##d, p = 0.0018 versus late non-RW/GT, F(2,11) = 4.96; two-way ANOVA, n = 4–6 session pairs. (A–H) Data are expressed as mean ± SEM.
(I) A model for the formation of the hippocampal salience map. The key processes in each phase are shown in red. During the early phase of map formation, de novo conversion from non-PCs primarily sets out a prototype map (left). Selective consolidation of GT cells and RW cells subsequently plays a dominant role in the establishment and maintenance of the salience map during the middle and late phases of training (right).
Since the analyses thus far considered active cells that exhibited detectable fluorescence changes, we further investigated the dynamics of inactive “silent” cells during the middle and late phases of map formation (for details, see STAR Methods). This independent analysis replicated the findings of selective stabilization of RW cells and GT cells and unbiased recruitment of non-RW/GT cells to these cell categories during map formation (recurrence of GT cells, 68.1% ± 7.0% [4.5-fold relative to a uniform distribution]; recurrence of RW cells, 50.8% ± 7.5% [2.5-fold relative to a uniform distribution]; recruitment of non-RW/GT cells to RW cells, 13.0% ± 3.2% [0.9-fold relative to a uniform distribution]; recruitment of non-RW/GT cells to GT cells, 14.2% ± 2.2% [0.7-fold relative to a uniform distribution]; n = 10 sessions of 893–1,219 cells from two mice). The analysis of silent cell dynamics revealed that the majority of silent cells (65.7% ± 2.2%, mean ± SD, n = 10 sessions of 1,576–1,682 cells from two mice) remained silent cells in the subsequent sessions. Among the silent cells that exited the pool of silent cells, 29.9% ± 4.6% and 70.1% ± 4.6% of them became PCs and non-PCs, respectively, while 29.9% ± 4.8% and 70.1% ± 4.8% of active-cell-derived silent cells returned from PCs and non-PCs, respectively. Of those silent-cell-derived PCs, 24.1% ± 4.8% (2.0-fold representation versus non-RW/GT cells) and 22.4% ± 4.5% (1.4-fold versus non-RW/GT cells) became GT cells and RW cells, respectively, whereas 22.3% ± 3.2% (1.8-fold versus non-RW/GT cells) and 22.6% ± 3.6% (1.3-fold versus non-RW/GT cells) of PC-derived silent cells returned from GT cells and RW cells, respectively. These results demonstrate that salient-location-biased PC formation from silent cells is equilibrated by a similarly biased elimination, as in the case of non-PCs (Figures 5A–5C). Silent cells and non-PCs thus share a similar mode of dynamics when they convert to and revert from PCs.
Separable Reward and Landmark Coding Revealed by Shank2-Deficient Mice
To dissect the molecular mechanisms underlying the over-representation of salient locations, we examined mice deficient in Shank2, a gene that encodes a scaffold protein found at the excitatory postsynaptic density (PSD). These mice mimic the microdeletion of Shank2 identified in a case of ASD with mild intellectual disability (Berkel et al., 2010), and exhibit ASD-like behaviors, including impaired social interaction and hyperactivity, reduced N-methyl-D-aspartate (NMDA) receptor function, and impaired long-term potentiation and long-term depression at hippocampal CA1 synapses (Won et al., 2012), providing a unique opportunity to test whether the integrity of postsynaptic signaling and synaptic plasticity is necessary for hippocampal map formation and plasticity.
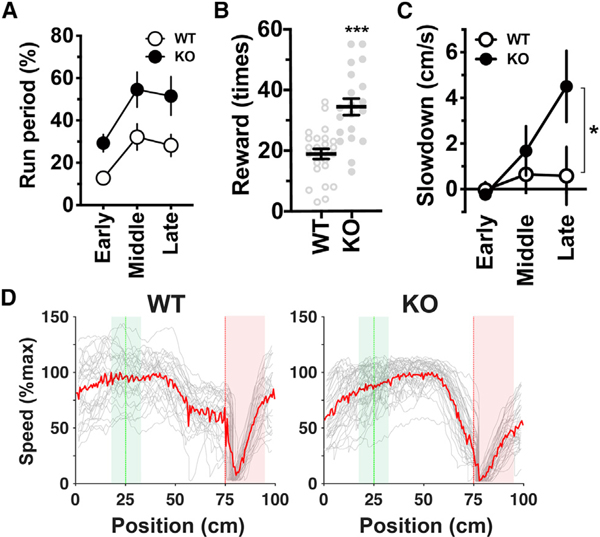
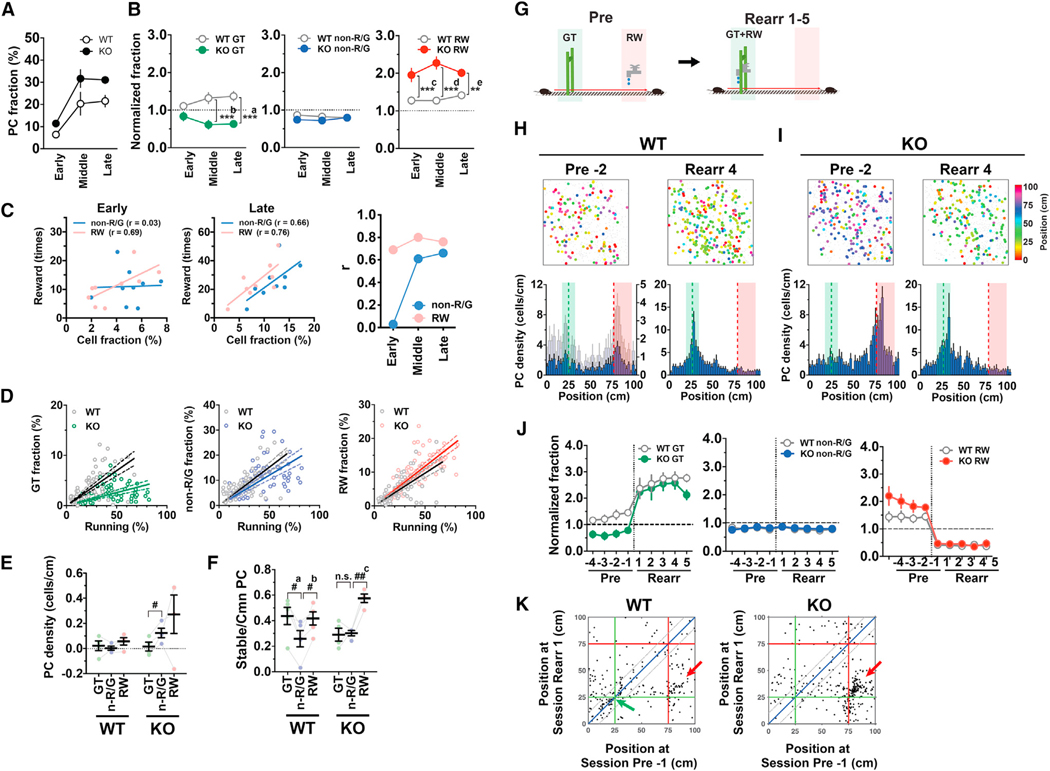
Shank2-deficient mice displayed not only an increased fraction of time spent running (Figure 6A; knockout [KO] versus wild type [WT], p = 0.025, F(1,7) = 8.14, two-way ANOVA, n = 5 WT and 4 KO mice) and acquisition of an increased number of rewards (Figure 6B) but also enhanced anticipatory slowdown before the reward zone (Figures 6C and 6D). This finding suggests that the hyperactivity of these mice is not merely a reckless run but rather reflects a more frequent repetition of highly stereotyped goal-directed behavior. However, the possibility that a higher level of attention and/or increased anxiety also contribute to this behavior cannot be excluded (Won et al., 2012). Consistent with the running-dependent increase in the fraction of PCs (Figure 2C), the overall PC fraction was elevated in Shank2-deficient mice compared to WT mice (Figure 7A; KO versus WT, p = 0.025, F(1,7) = 8.07, two-way ANOVA, n = 5 WT and 4 KO mice). Remarkably, the hippocampal maps of Shank2-deficient mice exhibited substantially enhanced over-representation of the reward location, whereas their learning-induced over-representation of the landmark location was selectively and completely lacking (Figures 7B, 7H, and 7I). It is unlikely that the absence of over-representation of the landmark location in Shank2-deficient mice was due to their potential impairment in vision, as they exhibited anticipatory behavior before the landmark even when the position of the landmark that indicated the reward location was randomly shifted (Figure S7). Compared to the average non-RW/GT cell fractions, the average RW cell fractions of each mouse had an increased correlation with the average number of rewards obtained throughout the training (Figure 7C), which indicates that RW cell fractions are a better indicator of the amount of rewarded experience. The fraction of GT cells for a given amount of spatial behavior was substantially reduced in Shank2-deficient mice, while those of non-RW/GT and RW cells were slightly decreased and increased, respectively (Figure 7D). Consistent with these findings, the net formation of GT cells from non-PCs during the early phase was significantly less than that of non-RW/GT cells in these mice (Figure 7E). Moreover, the stability of GT cells was reduced to a level comparable to that of non-RW/GT cells (Figure 7F). These findings indicate that Shank2 plays an indispensable role in both initial formation and subsequent stabilization of GT cells. Finally, we tested whether Shank2 is required for the reorganization of preformed maps in reward-rearrangement experiments, where mice trained on the standard virtual linear track for 15 sessions were further trained for the following five sessions on the same track with the exception that the location of reward was shifted to match the landmark location (Figure 7G). In contrast to the pivotal role of Shank2 in synaptic plasticity, the peak of the density of RW cells rapidly shifted to the new reward location in Shank2-deficient mice as in WT mice (Figures 7H–7J); thus, Shank2 is dispensable for reward-relocation-induced plasticity of RW cells (Figure 7K). In summary, the results clearly demonstrate that the two forms of hippocampal over-representation are mechanistically separable. Shank2 is required for learning-induced over-representation of landmark locations but not for rapid emergence and plasticity of over-representation of reward locations. The absence of Shank2 elicits abnormally enhanced over-representation of reward locations, and this “super-representation” is associated with enhanced running and goal-anticipation behavior and increased acquisition of rewarded outcomes.
Figure 6. Enhanced Goal-Directed Behavior of Shank2-Deficient Mice.
(A) The fraction of time spent running for the early (sessions 1–5), middle (sessions 6–10), and late (sessions 11–15) phases of training in wild-type (WT) and Shank2-deficient (KO) mice (WT versus KO, p = 0.025, F(1,7) = 8.14, two-way ANOVA, n = 5 WT and 4 KO mice).
(B) The number of rewards obtained in sessions in the late training phase. ***p < 0.0001 versus WT, t(43) = 5.07; unpaired two-tailed t test, n = 25 and 20 sessions from 5 WT and 4 KO mice, respectively.
(C) Slowdown before the reward delivery point *p = 0.031, F(1, 7) = 2.199, t(21) = 2.811, two-way ANOVA with the Holm-Sidak test, n = 5 WT and 4 KO mice. (A–C) Data are expressed as mean ± SEM.
(D) Examples of trial-by-trial (gray) and average (red) running speed plotted against position for WT (left) and KO (right) mice in single sessions in the late training phase.
Figure 7. Loss of Over-Representation of the Landmark Location and Augmented Over-Representation of the Reward Location in Shank2-Deficient Mice.
(A) PC fractions in the early, middle, and late phases of training (n = 5 WT and 4 KO mice).
(B) Normalized fractions of GT (left), non-RW/GT (middle), and RW (right) cells relative to the number of total PCs in each phase of training. ***a, p = 0.0009, ***b,p < 0.0001, F(1,7) = 22.39; ***c, p = 0.0014, **d, p = 0.0003, **e, p = 0.0003, F(1,7) = 33.33; two-way ANOVA, n = 5 WT and 4 KO mice.
(C) Correlations between the number of rewards obtained and the fractions of RW or non-RW/GT cells. The data for the early (average of sessions 1–5) and late (average of sessions 11–15) phases of training are shown in the left and middle panels, respectively. Each point represents the data from an individual animal, and the points and regression lines shown in red and blue represent those for RW and non-RW/GT cells, respectively. The graph shown right indicates the correlation coefficients of linear regressions for RW (red) and non-RW/GT (blue) cell fractions in the early, middle, and late phases of training. n = 5 WT and 4 KO mice.
(D) Scatterplots showing the relationship between the fraction of GT (left), non-RW/GT (middle), and RW (right) cells with respect to total identified cells and the fraction of time spent running. Each symbol represents an individual session. Solid and dotted lines indicate linear regression and 95% confidence intervals, respectively.
(E) Net formation of GT, non-RW/GT (n-R/G), and RW cells during the early training phase in WT and KO mice. #p = 0.046, F(1.025,3.075) = 2.35, t(3) = 4.979; one-way ANOVA with the Holm-Sidak test, n = 4 KO mice.
(F) The average PC stability for GT, non-RW/GT, and RW cells in WT and KO mice. #a, p = 0.036, #b, p = 0.034, F(1.938,7.753) = 11.36, one-way ANOVA, n = 5 WT mice. ##c, p = 0.0011, F(1.04,3.12) = 16.5, one-way ANOVA, n = 4 KO mice.
(G) Design of the reward-rearrangement task. Mice were first trained on the standard linear track, which included a visual landmark (GT) and reward delivery (RW) at separate locations, in preceding control sessions (pre). Once training was complete, the location of reward delivery was shifted to match the location of the landmark (GT+RW), and the mice were retrained in this new arrangement for the following five sessions (rearr 1–5).
(H and I) Example PC maps (top) and histograms indicating the average distribution of PCs with respect to position (bottom) before (pre ‒2) and after (rearr 4) reward rearrangement of WT (H) and KO mice (I).
(J) Normalized representations for GT (left), non-RW/GT (middle), and RW (right) cells in each session of reward-rearrangement experiments (n = 5 WT and 4 KO mice).
(K) Two-dimensional plots showing the positions of place fields of common PCs immediately before (pre ‒1) and after (rearr 1) the reward rearrangement in WT (left, n = 259 cells from five mice) and KO mice (right, n = 339 cells from four mice). Red arrows indicate clusters of cells that persistently encode the reward locations. The green arrow indicates a cluster of stable GT cells in WT mice.
(A), (B), (E), (F), and (H)–(J) Data are expressed as mean ± SEM.
DISCUSSION
In this study, we found that hippocampal over-representations of locations associated with reward and a visual landmark are mediated by separable subsets of neurons with different time courses of emergence and different underlying molecular mechanisms. The disproportionate hippocampal maps were established as a result of de novo conversion of the cells that encode each salient location from nonspatially tuned cells in the early phase, followed by their selective stabilization in the late phase. The two types of hippocampal over-representations were distinguishable by the necessity of Shank2; the learning-induced over-representation of the landmark location is dependent on Shank2, whereas rapid emergence and plasticity of over-representation of the reward location is independent of Shank2. Moreover, the abnormally distorted representation toward the reward location in the Shank2-deficient mouse model of ASD was associated with enhanced running and goal-anticipation behavior and increased acquisition of the rewarded experience, which suggests that aberrant hippocampal mapping of salient features may underlie cognitive and behavioral abnormalities in a subset of ASD cases.
In contrast to the preceding studies that examined only a single type of salient feature (O’Keefe and Conway, 1978; Wiener et al., 1989; Hetherington and Shapiro, 1997; Hollup et al., 2001; Dupret et al., 2010; Danielson et al., 2016; Geiller et al., 2017; Zaremba et al., 2017; Gauthier and Tank, 2018, Bourboulou et al., 2019), our study investigated the impacts of reward and a landmark separately side by side and provided strong evidence that multiple mechanistically distinct over-representations encode different types of salient features within a single hippocampal map. In our model, the number or fraction of a particular functional subset of neurons is determined by equilibrium between their stability and instability. During the early phase of map formation, new conversion of PCs from non-PCs creates a sparse prototype map in which “seed” GT cells and RW cells are laid out. The higher stability of each of these cell categories shifts the equilibrium upward to an elevated steady-state level and enables induction or maintenance of increased numbers of cells during the later phases. Representations of salient locations by these subsets of neurons constitute more stable singularities within hippocampal maps and may provide substrates by which different kinds of salient experience form lasting memory traces. Immediate emergence of over-representation for motivational salience is favorable for the rapid location of a source of positive reinforcement within the subject’s environment, whereas gradual mapping of environmental salience presumably reflects experience-dependent learning of the environment that could contribute to some form of landmark-based navigation (Sato et al., 2017; Le Merre et al., 2018). Thus, the multiplicity of functionally distinct salience coding revealed in our study expands the capacity of hippocampal maps to encode the presence and absence of a variety of nonspatial features onto their spatial representations.
We identified Shank2 as a single gene that is essential for the establishment of hippocampal over-representation. Since the global KO of Shank2 is thought to cause many changes throughout the brain, the relevant circuit mechanisms need to be elucidated in future studies. One possibility is that since the Shank2-deficient mice used in our study show regular synapse numbers yet impaired NMDA receptor function and synaptic plasticity in the hippocampal CA1 (Won et al., 2012), this suggests that normal NMDA receptor-mediated signaling fulfilled by Shank2 may play a role in the formation and stabilization of GT cells. Detailed circuit and molecular dissection of pathways that convey distinct salience signals for reward and landmark also remains to be conducted. Neurons in the lateral entorhinal cortex fire in the vicinity of objects when they are introduced into the environment (Deshmukh and Knierim, 2011; Tsao et al., 2013; Basu et al., 2016; Wang et al., 2018), which makes them a putative source of the salience signal for landmarks. The selective loss of landmark over-representation unveiled in the present study might be implicated in a specific type of navigation impairment in humans, landmark agnosia, which involves the temporal lobe and hippocampus as its major neural correlates (van der Ham et al., 2017), and could also shed light on spatial behavior in ASD (Smith, 2015). Salience signals may modulate PC stability via not only an NMDA-receptor-dependent mechanism but also a dopamine-dependent mechanism (Kentros et al., 1998, 2004). Loss of Shank2 did not abolish the rapid emergence and plasticity of RW cells. While a mouse model of 22q11.2 deletion syndrome that affects more than 20 genes shows deficits in hippocampal reward over-representation and goal-oriented learning (Zaremba et al., 2017), a specific deletion of the single Shank2 gene in our study leads to substantially augmented reward over-representation and enhanced running and goal-anticipation behavior, demonstrating that hippocampal reward representation and related behavior can be altered bidirectionally in mutually opposite directions in different mouse models of neurodevelopmental disorders. The appearance of reward over-representation preceding the emergence of anticipatory behavior during training raises the possibility that the former could play a role in the occurrence of the latter. Hypermotivated behavior and increased ventral striatal function have been reported in other Shank2-deficient rodent models (Pappas et al., 2017; Modi et al., 2018), suggesting a hypothesis that enhanced hippocampal reward mapping in the absence of Shank2 contributes to the development of augmented goal-anticipation behavior via increased interaction with striatal motivational circuits (Lansink et al., 2009).
STAR ★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Masaki Sato (masaaki.sato@riken.jp).
Materials Availability
The mouse line generated in this study is available from the RIKEN BioResource Center (http://www.brc.riken.jp/lab/animal/en/; stock number RBRC06579).
Data and Code Availability
The datasets and custom computer code associated with this study will be made available by the authors upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were conducted in accordance with institutional guidelines and protocols approved by the RIKEN and Kyoto University Animal Experiments Committees. Adult male Thy1-G-CaMP7 transgenic mice and those lacking Shank2 were used for the experiments. Homozygous Shank2-deficient mice (Won et al., 2012) carrying the Thy1-G-CaMP7 transgene and age- and gender-matched wild-type control transgenic mice were obtained by crossing male homozygous Thy1-G-CaMP7 transgenic, heterozygous Shank2-deficient mice with female heterozygous Shank2-deficient mice lacking the Thy1-G-CaMP7 transgene. All mice used were at least 12 weeks old and weighed 28–35 g at the beginning of surgery. The mice were housed in groups of one to four per cage on a 12 h-12 h light-dark cycle (with lights on at 6 pm and off at 6 am the next day). Thy1-G-CaMP7 mice were genotyped by PCR using the primers 5′-CTGCTGCCCGACAACCA-3′ and 5′-GTCGTCCTTGAAGAAGATGG-3’, which provided a 465-bp product of the G-CaMP7 coding sequence from tail DNA samples of transgene-positive mice. The wild-type and Shank2-deficient alleles were detected by PCR using the primers WT fwd 5′-GCTAGCATGACGTGTGTTGTG-3′ and rev 5′-ACCTGTGTGTGATTTCTGAC-3′, and the primers KO fwd2 5′-CCGACTGCATCTGCGTGTTC-3′ and rev 5′-ACCTGTGTGTGATTTCTGAC-3′, respectively (Won et al., 2012).
METHOD DETAILS
Generation of Thy1-G-CaMP7 transgenic mice
The cDNA encoding G-CaMP7 (Ohkura et al., 2012) ligated to the coding sequence of DsRed2 via a Thosea asigna virus-derived 2A peptide (T2A) sequence (Sato et al., 2015) was subcloned into the Xho I site of the modified mouse Thy1.2 promoter vector (Feng et al., 2000). The 8.7-kb DNA fragment was prepared by digestion with Not I and Pvu I restriction enzymes and subsequent gel purification and injected into the pronuclei of 466 fertilized eggs of C57BL/6J mice. From 32 offspring, 9 mice were identified as transgene positive, and 6 exhibited transgene expression in the brain. One founder mouse that expressed the transgene at a high level in the hippocampus was used for this study.
Analysis of transgene expression
Analysis of transgene expression in Thy1-G-CaMP7 mice was conducted essentially as described previously (Sato et al., 2015). The primary antibodies used in immunolabeling were rabbit anti-calbindin D-28K (1:500, AB1778, Millipore, Billerica, MA), rabbit anti-GAD65/67 (1:500, AB1511, Millipore), mouse anti-parvalbumin (1:1000, clone PARV-19, P3088, Sigma, St. Louis, MO), mouse anti-somatostatin (1:200, clone SOM-018, GTX71935, Gene Tex, Irvine, CA), rabbit anti-glial fibrillary acidic protein (GFAP) (1:1000, N1506, Dako, Denmark) and rabbit anti-Iba1 (1:1000, 019–19741, Wako Pure Chemical Industries, Ltd., Japan), and the secondary antibodies used were Alexa Fluor 647-labeled goat anti-rabbit and anti-mouse IgG antibodies (1:700–1000, A-21245 and A-21236, Thermo Fisher Scientific, Waltham, MA). Nuclear counterstaining was conducted in PBS containing 10 mg/ml Hoechst 33258 (Calbiochem) and 0.1% Triton X-100 at room temperature for 5 min. The densities of CA1 pyramidal cells were determined in the pyramidal cell layer of the dorsal CA1 hippocampus in 4 fields per animal (field size, 212 × 212 μm) and expressed as cell number per 200 μm length of the pyramidal cell layer. A quantitative analysis of G-CaMP7 and calbindin immunolabeling fluorescence signals was performed by averaging fluorescence intensities across cell bodies and normalizing them to those of the brightest cells within the field of view. High-calbindin cells were defined as those whose normalized calbindin signals were greater than or equal to 0.2, and the remaining cells were grouped as low-calbindin cells.
The histological analyses confirmed that the long-term transgenic expression of G-CaMP7 in Thy1-G-CaMP7 mice does not cause discernible toxicity. The pyramidal cell densities and overall distribution patterns of glial fibrillary acidic protein (GFAP)-positive astrocytes and Iba1-positive microglia were indistinguishable between Thy1-G-CaMP7 and wild-type (WT) C57BL/6 mice (Figure S1C; pyramidal cell density, 51.3 ± 2.4 cells/200 μm pyramidal cell layer in Thy1-G-CaMP7 mice versus 48.7 ± 1.2 cells/200 μm pyramidal cell layer in WT mice; t(14) = 0.975, p = 0.35, n = 8 fields each from 2 Thy1-G-CaMP7 and 2 WT mice, unpaired two-tailed t test). It was also found that in addition to strong hippocampal expression (Figure S1D), Thy1-G-CaMP7 mice express G-CaMP7 in diverse brain areas, including the cerebral cortex, olfactory bulb, brainstem and cerebellum (Figures S1E–S1P).
Surgery
Mice were anesthetized with isoflurane in ambient air (3% induction, 1.5% maintenance) and placed in a custom-made stereotaxic frame. To reduce secretions and brain edema, we administered atropine (0.3 mg/kg, s.c.) and dexamethasone (2 mg/kg, s.c.) prior to anesthesia. A circular piece of the scalp was removed, and the underlying bone was cleaned and dried. Three small screws were then placed in the skull (two at the suture of the interparietal and occipital bones and one on the right frontal bone) to provide anchors for the head plate. A thin layer of cyanoacrylate was applied to provide a substrate to which the dental acrylic could adhere.
A stainless steel head plate (25 mm length, 4 mm width, 1 mm thickness) with a wide circular opening (7 mm inner diameter and 10 mm outer diameter, the center is 2.5 mm off relative to the middle of the long side of the plate) was affixed to the skull using dental cement. The center of the opening was targeted at 2 mm posterior to the bregma and 2 mm lateral to the midline in the left hemisphere. The cement was mixed with black ink to block light entry from the LCD monitor into the microscope and placed onto the skull such that it covered the entire skull, including the anchor screws, except for the area of skull inside the opening of the head plate.
Optical window preparation was performed as described previously with modifications (Dombeck et al., 2010; Sato et al., 2016). A few days after the head plate surgery, a 2.5-mm-diameter circular craniotomy was created on the skull overlying the dorsal hippocampus. The dura was removed with forceps, and the overlying cortex was aspirated in a small amount at a time using a blunted 25-gauge needle connected to a vacuum pump. This step was continued with occasional irrigation with cortex buffer (123 mM NaCl, 5 mM KCl, 10 mM glucose, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, pH 7.4) until the white matter, including the corpus callosum, was exposed. Then, the top-most layers of the white matter were gently peeled aside by holding with the vacuum-connected blunted needle such that its minimal thickness remained covering the dorsal surface of the hippocampus. To minimize bleeding, aspiration was initiated from a cortical area devoid of large vessels, and bleeding was treated immediately with a piece of gelatin sponge (Spongel, Astellas Pharma, Tokyo, Japan) wetted with cortex buffer. An imaging window was then inserted to mechanically support the cranial hole, its surrounding tissue and the hippocampal surface. The imaging window consisted of a stainless steel ring (2.5 mm outer diameter, 2.2 mm inner diameter and 1.0 mm height) with a round coverslip (2.5 mm diameter, 0.17 mm thickness, Matsunami Glass Ind., Osaka, Japan) attached to the bottom using a UV-curable adhesive (NOA81, Norland Products, Cranbury, NJ). To reduce brain movement during imaging, a small disk of medical grade clear silicone sheeting (0.13 mm thickness, 20–10685, Invotec International, Jacksonville, FL) was attached to the surface of the coverslip facing the hippocampal tissue (Mower et al., 2011). When the window was positioned, the bottom coverslip was approximately parallel relative to the head plate, and the hippocampal surface was clearly visible through the bottom coverslip without any trace of bleeding. The upper rim was then cemented to the skull with dental acrylic.
After surgery, a metal cover (0.3 mm thickness) was screwed onto the upper surface of the head plate to protect the imaging window from dust. The mice were placed in a warmed chamber until they fully recovered from anesthesia and were then returned to their home cages. They were housed for at least 4 weeks of postoperative recovery before the start of handling.
VR set-up
A VR system with an air-supported spherical treadmill for head-fixed mice was constructed as described previously (Sato et al., 2017). A 20-cm-diameter Styrofoam ball placed inside the bowl provided a freely rotating surface on which the mouse stood. The mouse was positioned near the top of the ball with its head fixed via the steel head plate that was screwed into a rigid cross bar and posts. A single wide-screen 23” LCD display (Dell U2312, Round Rock, TX) placed 30 cm in front of the mice presented VR scenes rendered by OmegaSpace 3.1 (Solidray Co. Ltd., Yokohama, Japan) running on a Windows 7 computer in 81° horizontal and 51° vertical fields of view. The LCD monitor was large enough to cover the major part of the mouse’s binocular and monocular visual fields (Sato and Stryker, 2008). The use of a single LCD monitor for VR presentation effectively elicits visual cue-based virtual navigation behavior in head-fixed mice (Youngstrom and Strowbridge, 2012; Sato et al., 2017).
The movement of the ball was measured with a USB optical computer mouse (G400, Logitech, Newark, CA) via custom driver and LabVIEW software (National Instruments, Austin, TX). The optical mouse was positioned in front of the mouse and at the intersection of the mouse’s sagittal plane and the equator of the ball. The signals along the horizontal axis (aligned parallel to the mouse’s sagittal plane) generated by the running of the head-fixed mouse was used to compute rotational velocity in the forward-backward direction. This velocity signal was converted into analog control voltages (0–5 V) via a D/A converter and fed to a USB joystick controller (BU0836X, Leo Bodner, Northamptonshire, UK) connected to the OmegaSpace computer to move the mouse’s position in VR.
Water rewards (5 μl/reward) were delivered by a microdispenser unit (O’Hara & Co., Ltd., Tokyo, Japan) attached to a waterfeeding tube positioned directly in front of the mouse’s mouth. The unit was triggered upon reward events in VR by 5 V TTL signals generated by an OmegaSpace script via a USB-connected D/A device (USB-6009, National Instruments). The behavioral parameters, such as the mouse’s location in the virtual environment, the trigger signals for water rewards and the rotational velocity signals of the spherical treadmill, were recorded at 20-ms intervals using custom software in LabVIEW. The TTL signals for each frame sent by the microscope computer were recorded with the behavioral data to synchronize the imaging and behavioral data.
Behavior
At least 5 days before the start of imaging experiments, mice implanted with the head plate and the imaging window were acclimated to handling and the Styrofoam ball. During this acclimation, mice were handled by an experimenter for 5–10 min and then allowed to move freely on the top of the ball, which was rotated manually by the experimenter, for another 5–10 min. The procedure was performed once a day and repeated for at least 3 days. The mice were then placed on a water restriction schedule 2–3 days before the start of the experiments. No other type of pre-training was conducted before the mice were exposed to the virtual environment in the first sessions. Body weight and general appearance were checked daily to ensure that the animals maintained at least ~85% of their preoperative body weight and exhibited no signs of abnormal behavior throughout the study. The experiments were performed during the dark phase of the cycle to enhance the locomotion of the mice.
The virtual endless linear track was created using an editor function in OmegaSpace. The mouse started at the origin of the virtual linear track segment and ran along the track unidirectionally with visual feedback rendered by OmegaSpace. The track segment was 100 cm long, measured as the number of rotations of the ball required to move from one end of the track to the other times the circumference of the ball. The mouse moved only one-dimensionally along the midline of the track, with its view angle fixed in the direction of movement. Different patterns were placed on the walls of each track subsegment as follows: vertical white and black stripes at 0–25 cm, horizontal white and black stripes at 25–50 cm, and black dots on a white background at 50–100 cm. The floor was patterned with a white grid on a black background. The space above the track was colored black. A green gate was placed 25 cm from the origin to serve as a salient landmark. Water rewards were delivered when the mouse reached a reward point located 75 cm from the origin. This reward point was located in the middle of a track zone with a certain wall pattern (i.e., black dots on a white background) and not denoted with any other salient visual cues. Upon reaching the other end of the segment, the mouse’s virtual position was transferred back to the origin, and the same segment of the linear track was presented again. The approaching track segment following the current segment was always rendered on the monitor, such that the virtual linear track appeared infinitely long.
The mice underwent a total of 15 training sessions in the above task, with 1–2 sessions per day. Each session was 10 min long. When 2 sessions were performed in one day, the intervals between sessions were at least 4 h, and the mice were returned to their home cages between the sessions. The entire training period from the first to the last session was 225 ± 8h (mean ± SD, n = 7 mice). Each mouse was lightly anesthetized with isoflurane to detach the metal window cover screwed onto the head plate and clean the imaging window, after which the mouse was placed into the VR apparatus. The head was then fixed to the crossbar above the ball via the head plate, and the mouse was left on the ball in the dark for approximately 20 min until it had recovered fully from the anesthesia. During the behavioral session, the animal was allowed to behave freely in the head-fixed arrangement. G-CaMP7 fluorescence in hippocampal CA1 pyramidal neurons was simultaneously imaged as described below. Licking was detected using an infrared photo beam sensor (OPR-LKR, O’Hara & Co., Ltd.) in a subset of experiments. Slowdown of running speed before the reward delivery point was calculated as the difference between the maximum average running speed between the landmark location and the reward delivery point (32.5–65 cm from the origin) and the average running speed in the area immediately before the reward delivery point (65–75 cm from the origin).
For the reward-rearrangement task, mice first underwent 15 training sessions on the virtual linear track as described above. The mice were further trained for the following 5 sessions (Rearrangement 1–5) in the same virtual linear track except that the location of reward delivery (75 cm from the origin) was shifted to match the location of the landmark (25 cm from the origin). Data obtained from the last 4 sessions of the initial 15 training sessions before the shift (Sessions 12 through 15, also referred to as Pre −4 through −1) were analyzed as pre-rearrangement baseline sessions. The first rearrangement sessions were performed immediately after the last baseline sessions without releasing the mice from head fixation.
For the task with 50% reward delivery and the task with no gate, a separate cohort of mice (n = 3) were trained in 15 normal sessions and were then subjected to 2 sessions of the 50% reward task, in which the reward was delivered at the same delivery point with 50% probability, followed by 2 normal sessions and 1 no-gate session, where the green gate was removed from its location throughout the entire sessions. The first 50% reward sessions and the no-gate sessions were performed immediately after the preceding normal sessions without releasing the mice from head fixation.
For the random gate shift task, mice (n = 3 wild-type mice and 3 Shank2-deficient mice) were first trained on the virtual linear track in which the visual landmark indicated the reward location at 62.5 cm from the origin for 15 sessions. Then, each of the following 5 random shift sessions began with 12 baseline trials in which the position of the landmark plus reward was fixed at the pre-trained location, followed by random shift trials in which their position was shifted forward or backward by 12.5 cm or remained at the pre-trained (normal) position on a trial-by-trial basis in a random manner. Each of the three landmark plus reward position was tested four times for a total of 12 random shift trials. Slowdown and licking immediately (0–10 cm) before the landmark plus reward location were analyzed separately for baseline, forward, normal and backward trials in each session and averaged across the last three of the five sessions for each mouse.
Imaging
Imaging was performed using a Nikon A1MP (Nikon, Tokyo, Japan) equipped with a 16x, NA 0.8 water immersion objective lens. The microscope was controlled with Nikon NIS-elements software. G-CaMP7 and DsRed2 were excited using a Ti-sapphire laser (MaiTai DeepSee eHP, Spectra-Physics, Santa Clara, CA) at 910 nm. Typical laser power was approximately 40 mW at the objective lens. G-CaMP7 fluorescence was separated using a 560-nm dichroic mirror and collected with an external GaAsP photomultiplier tube (10770PB-40, Hamamatsu Photonics, Hamamatsu, Japan) mounted immediately above the objective lens. The calcium-insensitive DsRed2 fluorescence, which helped to identify G-CaMP7-labeled pyramidal neurons, was simultaneously imaged and recorded using another GaAsP photomultiplier tube. The DsRed2 images were checked by the experimenter for the on-site assessment of the quality of image acquisition but not used for offline quantitative image analysis, except for image alignment across sessions (Figure S5).
To image G-CaMP7-labeled CA1 pyramidal neurons, the microscope was focused at a depth of approximately 150 μm from the hippocampal surface. To prevent the entry of light from the LCD monitor into the microscope, a small sheet of aluminum foil was wrapped around the objective lens, so the foil completely covered the space between the objective and the skull. Images of 512 × 512 pixels were acquired at a rate of 15 frames per second using a resonant-galvo scanner mounted on the microscope. Each imaging session was 10 min long. The size of the field of view was 532 × 532 μm. In repeated chronic imaging, previously imaged cell populations usually re-appeared at similar depths in new sessions. We took reference images of DsRed2 fluorescence at the beginning of each session to confirm that the reference image of the current session was very similar to that of the previous session by ensuring that blood vessels and neurons arranged in unique patterns appeared in the same parts of the two images.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
Each frame of a G-CaMP7 time-lapse movie was aligned to an average fluorescence image of the movie for motion correction using the TurboReg ImageJ plug-in. The registered movie was then denoised by a spatio-temporal median filter. This preprocessed movie f(t,x) was reconstituted to the sum of fluorescence intensity of individual cells using a modified non-negative matrix factorization algorithm, as described in detail elsewhere (Vogelstein et al., 2010; Pnevmatikakis et al., 2016; Takekawa et al., 2017; Giovannucci et al., 2019). Briefly, this algorithm assumes that the fluorescence intensity of each cell can be deconvoluted to the spatial filter ac(x), which represents the position and shape of the cell, and the time variation vc(t) derived from spiking activities uc(t):
where a0, v0 are baselines, and σ2 is intensity of Gaussian noise. As is the case in cell identification using independent component analysis (Mukamel et al., 2009), this algorithm preferentially detects cells that change their fluorescence intensities over time (“active cells”) because cells that barely do so are regarded as being near baseline. Each spike derives the transient elevation of fluorescence intensity with a double-exponential shape:
The exponential rise and decay time constants τ1 = 0.09 and τ2 = 0.261, respectively, were obtained by curve fitting of actual traces of cellular calcium transients in G-CaMP7-expressing CA1 pyramidal neurons in Thy1-G-CaMP7 mice in vivo. Spatial filters and spike timings were estimated by two iterative steps. In the first step, we prepared tentative spatial filters and estimated spike trains corresponding to respective filters by a least-squares approach with a non-negative restraint condition. Subsequently, spatial filters were estimated using the least-squares method on the condition that the estimated spike trains were feasible. In addition, we introduced L1 sparse regularization derived from priors that represented the typical cell size and spike frequency. To determine the mutual relationship between a and u, a regularized term was also introduced to the model. This condition guaranteed the uniqueness of the scale of a, u and v. As a consequence, a, u and v are presented in arbitrary units, while the product of a and v corresponds to the observed data.
In practice, 512 × 512 pixel image data were divided into 4 × 4 of 128 × 128 pixel subareas with 32-pixel overlap regions. Each subarea was analyzed with the above algorithm, and the results were combined to cover the whole image area. After the initial calculation, the morphology of each spatial filter was defined as the region above 0.2 times its peak value, and the position of the filter was defined by its weighted centroid. We then removed the following filters as those that did not represent complete single pyramidal cell morphology: (1) filters whose areas were smaller than 25 pixels, (2) filters whose areas were larger than 400 pixels, (3) filters located on the edge of the image, (4) filters whose heights or widths were greater than 64 pixels because they often contained structures of multiple cells, and (5) smaller filters in filter pairs whose distances were closer than 10 pixels (10.4 μm) and whose temporal correlation coefficients of activities were greater than 0.3 because they were considered to be derived from the same cell.
After those non-cell filters were removed, we recalculated the activity time series for the new filter set. Visual inspection confirmed that nearly all active cells that were represented in a background-subtracted maximum-intensity projection image were identified with this procedure (Figures S3A and S3B). All images of the entire session, regardless of the mouse’s behavioral state, were used for this image analysis. The average number of cells identified from a movie of a session was 900 ± 246 (mean ± SD, n = 105 sessions from 7 mice).
Analysis of place fields
Place fields were calculated using cellular activity during movement periods. We defined these periods as the time when the mouse moved at a speed of > 0.5 cm/s continuously for a duration of > 2 s to reject irrelevant movements, such as grooming and jittering on the ball. We divided the entire virtual linear track segment into 80 bins (bin size = 1.25 cm) and created a histogram of neuronal activity versus track position for each cell. The activity events were defined by binarizing the time series of inferred spike activity u at a threshold of 0.1, which was empirically determined to remove baseline noise. The counts on the histogram were then divided by the mouse’s occupancy time in each bin, and the resultant place fields were smoothed with a Gaussian function (Gaussian window size = 6.25 cm) and normalized to the maximum values. To test the significance of place-related activity, we calculated the mutual information content between neuronal activity and the mouse’s virtual location for each cell (Markus et al., 1994; Ziv et al., 2013). We compared this value to a distribution of mutual information content calculated using 1000 randomly permuted data for the same cell. The permutation was conducted by rotating the activity event time series by a random amount relative to the time series of the mouse’s virtual positions. Cells were considered place cells (PCs) if their overall activity rates within the session were no less than 0.1 events/s and their mutual information content in the real data was greater than the 95th percentile of the values obtained from the randomly permuted data. We defined the position of the place field of each PC by the position of the peak of the field. A PC was considered a “gate (GT) cell,” “reward (RW) cell” or “wall (WL) cell” if the position of its place field was 17.5–32.5, 75–95 or 47.5–55 cm from the origin of the track segment. PCs with place fields outside the above zones were categorized as “non-reward, non-gate cells (non-RW/GT cells).” The PC formation factor was defined by the slope of a least-squares regression line fitted to a plot of the fraction of PCs against the fraction of time spent running, which contained data points from all tested animals in the session of interest. The linear regression model included no constant term under the assumption that no PCs were formed without running in any given session. When we calculated the fractions of GT, RW and non-RW/GT cells relative to the number of total PCs, we used data from sessions with at least 35 total PCs to avoid the effects of improperly large or small fractions caused by small numbers of cells.
Analysis of cellular activity in the 50% reward task was conducted by dividing the data from the full sessions into two subsets, namely, rewarded trials and non-rewarded trials, and analyzing them separately. The activity bias index between rewarded trials and non-rewarded trials was then calculated as (R-N)/(R+N), where R and N represent the cell’s in-field activity in rewarded trials and non-rewarded trials, respectively. The activity bias index ranges from −1 to 1, where a positive value indicates a bias toward rewarded trials and a negative value indicates a bias toward non-rewarded trials. Cells that had activity bias indices < −0.1 (equivalent to an approximately 1.22-fold increase in non-rewarded trials), −0.1 ≤ activity bias indices < 0.1 (equivalent to an approximately 1.22-fold increase in rewarded trials), and activity bias indices ≥ 0.1 were subsequently categorized as no-reward-preferring RW cells, constant RW cells, and reward-preferring RW cells, respectively. In the analysis of cellular activity in the no-gate task, the same cells that were identified both in the preceding control (pre) and no-gate sessions were determined as described below (“Alignment of cells across sessions”), and the calculation of the activity bias index between pre and no-gate sessions and the categorization of gate-preferring GT cells, no-gate-preferring GT cells, and constant GT cells was conducted as in the 50% reward task.
Alignment of cells across sessions
To find a consistent population of cells in images that were acquired in two different sessions, we first estimated the extent of overall image displacement that existed between the two image datasets. We searched for a peak in the two-dimensional correlation co-efficient calculated between the two DsRed2 reference images obtained at the beginning of each session within a range of 25 × 25 pixels (26.0 × 26.0 μm) of displacement in the x and y dimensions (Figure S5A). In this analysis, we conservatively focused on comparisons between two consecutive sessions (i.e., sessions 1 and 2, 2 and 3, etc.) because the quality of the image alignment was gradually reduced as the number of sessions that separated the two images increased (p < 0.0001, F(13,721) = 10.94, one-way ANOVA, Figure S5F). A preliminary assessment of the first cohort confirmed that every compared image pair displayed a peak within this range (average displacement in the x dimension, 5.9 ± 4.5 μm; average displacement in the y dimension, 4.7 ± 4.2 μm; average peak correlation coefficient, 0.77 ± 0.09; mean ± SD; n = 98 image pairs). During the calculation of two-dimensional correlation coefficients, the image of one session (the “source” session) was systematically shifted relative to that of the other session (the “target” session). The map of the coordinates of all cell positions in the target session was then overlaid with that of the source session, shifted by the amount of the estimated displacement (Figure S5A). The cell closest to each cell in the target session was searched in the displaced source session map, and the cell that was found was regarded provisionally to be the same cell if they were separated by 5 pixels (5.2 μm) or less. Cells that were unable to find the closest cells within this range were rejected from the subsequent analysis. After finding the provisional counterparts in the displaced source session map, the same procedure was repeated for the cells in the displaced source session map to conversely find their closest partners in the target session. This step helped remove cell pairs that were redundantly assigned (e.g., two different cells in one session falsely assigned to the same single cell in the other session) and the resultant cell pairs that had mutually unique correspondence were considered to be the pairs that represented the same cells (termed hereafter “common cells”). When comparing PC maps, common PCs were defined as a subset of common cells that were identified as significant PCs in both consecutive sessions. Stable PCs were defined as a subset of common PCs with place-field positions in the consecutive sessions that were close to each other (i.e., place field distance < 10 cm). In the analysis of formation, recruitment and stabilization of PCs (Figure 5), we first identified a population of common cells that belonged to the cell category of interest in the reference session N and then tracked the position of the place field of each cell in the subsequent session N+1. The transition was quantified by determining the density of cells at each bin in session N+1.
Since the above-described pairwise cell matching can identify only the correspondence between active cells in adjacent sessions, a subset of data (n = 2 mice, 15 training sessions from each) was further analyzed including inactive (“silent”) cells as follows. Spatial filters and spike timings of all sessions were estimated from G-CaMP7 time-lapse movies with the constrained nonnegative matrix factorization (CNMF) implementation developed by Pnevmatikakis et al. (2016) and Giovannucci et al. (2019). Common spatial filters were detected using a model that finds the most probable counterpart by jointly taking into account their centroid distances and correlations between spatial filters (Sheintuch et al., 2017). This procedure found common spatial filters in 77% of filters examined (n = 32,816) in two adjacent sessions. This search was then extensively conducted in every pair within a series of sessions, and clusters of common spatial filters detected in at least two different sessions were selected as candidate cells. These cells were considered “silent” in the sessions in which no corresponding spatial filters were identified. We confirmed that none of these missing regions contained individual cell-like activity by calculating the probability of obtaining the maximum amplitude deviation from baseline activity under given baseline fluctuations. Map comparisons including these silent cells were conducted as described above except that they additionally took into account the transitions from and to silent cells.
Statistics
Statistical details including sample sizes can be found in the figure legends and Results. When only two groups were compared, two-sided Student’s t tests were used if the variances of the two groups were similar. Otherwise, two-tailed Mann-Whitney tests were used. When more than two groups were compared, analysis of variance (ANOVA) was used if variances of the groups compared were similar. Otherwise, a non-parametric version of ANOVA (Friedman test) was used. In both parametric and non-parametric ANOVA, p values were adjusted for post hoc multiple comparisons. Exact p values are shown unless p < 0.0001. Statistical tests were performed using GraphPad Prism versions 6 and 7 (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-calbindin D-128K | Millipore | Cat#AB1778; RRID: AB_2068336 |
| Rabbit anti-GAD65/67 | Millipore | Cat#AB1511; RRID: AB_90715 |
| Mouse anti-parvalbumin (clone PARV-19) | Sigma | Cat#P3088; RRID: AB_477329 |
| Mouse anti-somatostatin | GeneTex | Cat#GTX71935; RRID: AB_383280 |
| Rabbit anti-glial fibrillary acidic protein | Dako | Cat#N1506; RRID: AB_10013482 |
| Rabbit anti-Iba1 | Wako | Cat#019-19741; RRID: AB_839504 |
| Alexa Fluor 647-labeled goat anti-rabbit IgG | Thermo Fisher | Cat#A-21245; RRID: AB_2535813 |
| Alexa Fluor 647-labeled goat anti-mouse IgG | Thermo Fisher | Cat#A-21236; RRID: AB_2535805 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Thy1-G-CaMP: C57BL/6J-Tg(Thy1-G-CaMP7, -DsRed2)492Bsi | This paper | RIKEN BRC: RBRC06579 |
| Mouse: Shank2-deficient mice: B6N.129S4-Shank2tm1Mgle/CsbdJ | Won et al., 2012 | JAX: 033667; RRID: IMSR_JAX033667 |
| Oligonucleotides | ||
| Genotyping primer for Thy1-G-CaMP7 mice: CTGCTGCCCGACAACCA | Sato et al., 2015 | N/A |
| Genotyping primer for Thy1-G-CaMP7 mice: GTCGTCCTTGAAGAAGATGG | Sato et al., 2015 | N/A |
| Genotyping primer for Shank2-deficient mice: Shank2 WT fwd: GCTAGCATGACGTGTGTTGTG |
Won et al., 2012 | N/A |
| Genotyping primer for Shank2-deficient mice: Shank2 KO fwd2: CCGACTGCATCTGCGTGTTC |
Won et al., 2012 | N/A |
| Genotyping primer for Shank2-deficient mice: Shank2 rev: ACCTGTGTGTGATTTCTGAC | Won et al., 2012 | N/A |
| Recombinant DNA | ||
| Mouse Thy1.2 promoter | Feng et al., 2000 | RRID: Addgene_20736 |
| G-CaMP7-T2A-DsRed2 | Ohkura et al., 2012 Sato et al., 2015 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | RRID: SCR_003070 |
| TurboReg | Biomedical Imaging Group, Swiss Federal Institute of Technology Lausanne | RRID: SCR_014308 |
| MATLAB | Mathworks | RRID: SCR_001622 |
| LabVIEW | National Instruments | RRID: SCR_014325 |
| OmegaSpace | Solidray | http://www.solidray.co.jp/product/omega/index.html |
| Prism | GraphPad | RRID: SCR_002798 |
| Modified nonnegative matrix factorization | Takekawa et al., 2017 | https://www.biorxiv.org/content/10.1101/215145v1 |
| Constrained nonnegative matrix factorization |
Pnevmatikakis et al., 2016 Giovannucci et al., 2019 |
https://github.com/flatironinstitute/CaImAn |
| Cell registration | Sheintuch et al., 2017 | https://github.com/zivlab/CellReg |
Highlights.
CA1 over-representation of reward and landmark emerge with distinct time courses
These cells form stable singularities during experience-dependent map consolidation
The over-representation of landmark but not reward is dependent on Shank2
ACKNOWLEDGMENTS
We thank Charles Yokoyama, Thomas J. McHugh, Shigeyoshi Fujisawa, Liset Menendez de la Prida, and Hokto Kazama for helpful comments and discussion. This work was supported by Precursory Research for Embryonic Science and Technology (PRESTO) JPMJPR12A1 from the Japan Science and Technology Agency (JST), KAKENHI grants 24700403, 25116528, 26115530, 17H05985, 19H04942, and 20H03550 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS), and a grant from the Senshin Medical Research Foundation to M.S.; RIKEN, NIH grant R01DA17310, KAKENHI grants 22110006, 16H01292, and 18H05434, a Human Frontier Science Programme grant, and Fujitsu Laboratories to Y.H.; KAKENHI grants 25830023, 15H01571, 17H05695, and 19K16293 to K.M.; KAKENHI grant 26870577 to T.T.; KAKENHI grant 15H04265 to T.F; KAKENHI grants 26115504, 15H05723, and 16H06536, and the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from MEXT and the Japan Agency for Medical Research and Development (AMED) to J.N.; Regional Innovation Cluster Program grant (City Area Type, Central Saitama Area) from MEXT to J.N. and M.O.; and grants from the VW Foundation (ZN2632), BCCN (01GQ1005A and 01GQ1005B), DFG (CRC 1286, 889), the Ministry for Science and Culture of Lower Saxony, and the Max Planck Society to F.W. D.G.-D. is a recipient of the PhD fellowship BES-2013-064171 and a grant from the short-term visit program EEBB-I-15-09552.
DECLARATION OF INTERESTS
Y.H. was supported in part by Takeda Pharmaceutical and DWANGO.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107864.
REFERENCES
- Basu J, Zaremba JD, Cheung SK, Hitti FL, Zemelman BV, Losonczy A, and Siegelbaum SA (2016). Gating of hippocampal activity, plasticity, and memory by entorhinal cortex long-range inhibition. Science 351, aaa5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. (2010). Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet 42, 489–491. [DOI] [PubMed] [Google Scholar]
- Bostock E, Muller RU, and Kubie JL (1991). Experience-dependent modifications of hippocampal place cell firing. Hippocampus 1, 193–205. [DOI] [PubMed] [Google Scholar]
- Bourboulou R, Marti G, Michon FX, El Feghaly E, Nouguier M, Robbe D, Koenig J, and Epsztein J. (2019). Dynamic control of hippocampal spatial coding resolution by local visual cues. eLife 8, e44487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess N, and O’Keefe J. (1996). Neuronal computations underlying the firing of place cells and their role in navigation. Hippocampus 6, 749–762. [DOI] [PubMed] [Google Scholar]
- Chen G, King JA, Burgess N, and O’Keefe J. (2013). How vision and movement combine in the hippocampal place code. Proc. Natl. Acad. Sci. USA 110, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M, and Losonczy A. (2016). Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area CA1. Neuron 91, 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh SS, and Knierim JJ (2011). Representation of non-spatial and spatial information in the lateral entorhinal cortex. Front. Behav. Neurosci 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, and Tank DW (2010). Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci 13, 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupret D, O’Neill J, Pleydell-Bouverie B, and Csicsvari J. (2010). The reorganization and reactivation of hippocampal maps predict spatial memory performance. Nat. Neurosci 13, 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, and Cohen NJ (2014). Can we reconcile the declarative memory and spatial navigation views on hippocampal function? Neuron 83, 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, and Sanes JR (2000). Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51. [DOI] [PubMed] [Google Scholar]
- Frank LM, Stanley GB, and Brown EN (2004). Hippocampal plasticity across multiple days of exposure to novel environments. J. Neurosci 24, 7681–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JL, and Tank DW (2018). A dedicated population for reward coding in the hippocampus. Neuron 99, 179–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiller T, Fattahi M, Choi JS, and Royer S. (2017). Place cells are more strongly tied to landmarks in deep than in superficial CA1. Nat. Commun 8, 14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P, et al. (2019). CaImAn an open source tool for scalable calcium imaging data analysis. eLife 8, e38173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, Skaggs WE, Moore KM, and McNaughton BL (1996). Binding of hippocampal CA1 neural activity to multiple reference frames in a landmark-based navigation task. J. Neurosci 16, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington PA, and Shapiro ML (1997). Hippocampal place fields are altered by the removal of single visual cues in a distance-dependent manner. Behav. Neurosci 111, 20–34. [DOI] [PubMed] [Google Scholar]
- Hill AJ (1978). First occurrence of hippocampal spatial firing in a new environment. Exp. Neurol 62, 282–297. [DOI] [PubMed] [Google Scholar]
- Hollup SA, Molden S, Donnett JG, Moser MB, and Moser EI (2001). Accumulation of hippocampal place fields at the goal location in an annular watermaze task. J. Neurosci 21, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentros C, Hargreaves E, Hawkins RD, Kandel ER, Shapiro M, and Muller RV (1998). Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science 280, 2121–2126. [DOI] [PubMed] [Google Scholar]
- Kentros CG, Agnihotri NT, Streater S, Hawkins RD, and Kandel ER (2004). Increased attention to spatial context increases both place field stability and spatial memory. Neuron 42, 283–295. [DOI] [PubMed] [Google Scholar]
- Kohara K, Pignatelli M, Rivest AJ, Jung HY, Kitamura T, Suh J, Frank D, Kajikawa K, Mise N, Obata Y, et al. (2014). Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci 17, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komorowski RW, Manns JR, and Eichenbaum H. (2009). Robust conjunctive item-place coding by hippocampal neurons parallels learning what happens where. J. Neurosci 29, 9918–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansink CS, Goltstein PM, Lankelma JV, McNaughton BL, and Pennartz CMA (2009). Hippocampus leads ventral striatum in replay of placereward information. PLoS Biol. 7, e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merre P, Esmaeili V, Charrière E, Galan K, Salin PA, Petersen CCH, and Crochet S. (2018). Reward-based learning drives rapid sensory signals in medial prefrontal cortex and dorsal hippocampus necessary for goal-directed behavior. Neuron 97, 83–91.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Marchionni I, Bezaire M, Varga C, Danielson N, Lovett-Barron M, Losonczy A, and Soltesz I. (2014). Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron 82, 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Treves A, Meyer R, Barnes CA, McNaughton BL, Moser MB, and Moser EI (2005). Progressive transformation of hippocampal neuronal representations in “morphed” environments. Neuron 48, 345–358. [DOI] [PubMed] [Google Scholar]
- Lever C, Wills T, Cacucci F, Burgess N, and O’Keefe J. (2002). Long-term plasticity in hippocampal place-cell representation of environmental geometry. Nature 416, 90–94. [DOI] [PubMed] [Google Scholar]
- Markus EJ, Barnes CA, McNaughton BL, Gladden VL, and Skaggs WE (1994). Spatial information content and reliability of hippocampal CA1 neurons: effects of visual input. Hippocampus 4, 410–421. [DOI] [PubMed] [Google Scholar]
- Mehta MR, Barnes CA, and McNaughton BL (1997). Experience-dependent, asymmetric expansion of hippocampal place fields. Proc. Natl. Acad. Sci. USA 94, 8918–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K, Diba K, Pastalkova E, and Buzsáki G. (2011). Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nat. Neurosci 14, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi ME, Brooks JM, Guilmette ER, Beyna M, Graf R, Reim D, Schmeisser MJ, Boeckers TM, O’Donnell P, and Buhl DL (2018). Hyperactivity and hypermotivation associated with increased striatal mGluR1 signaling in a Shank2 rat model of autism. Front. Mol. Neurosci 11, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower AF, Kwok S, Yu H, Majewska AK, Okamoto K, Hayashi Y, and Sur M. (2011). Experience-dependent regulation of CaMKII activity within single visual cortex synapses in vivo. Proc. Natl. Acad. Sci. USA 108, 21241–21246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, and Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RU, and Kubie JL (1987). The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J. Neurosci 7, 1951–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe J, and Conway DH (1978). Hippocampal place units in the freely moving rat: why they fire where they fire. Exp. Brain Res 31, 573–590. [DOI] [PubMed] [Google Scholar]
- O’Keefe J, and Nadel L. (1978). The Hippocampus as a Cognitive Map (Clarendon; ). [Google Scholar]
- Ohkura M, Sasaki T, Sadakari J, Gengyo-Ando K, Kagawa-Nagamura Y, Kobayashi C, Ikegaya Y, and Nakai J. (2012). Genetically encoded green fluorescent Ca2+ indicators with improved detectability for neuronal Ca2+ signals. PLoS ONE 7, e51286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas AL, Bey AL, Wang X, Rossi M, Kim YH, Yan H, Porkka F, Duffney LJ, Phillips SM, Cao X, et al. (2017). Deficiency of Shank2 causes mania-like behavior that responds to mood stabilizers. JCI Insight 2, e92052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, and Stryker MP (2008). Distinctive features of adult ocular dominance plasticity. J. Neurosci 28, 10278–10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Kawano M, Ohkura M, Gengyo-Ando K, Nakai J, and Hayashi Y. (2015). Generation and imaging of transgenic mice that express G-CaMP7 under a tetracycline response element. PLoS ONE 10, e0125354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Kawano M, Yanagawa Y, and Hayashi Y. (2016). In vivo two-photon imaging of striatal neuronal circuits in mice. Neurobiol. Learn. Mem 135, 146–151. [DOI] [PubMed] [Google Scholar]
- Sato M, Kawano M, Mizuta K, Islam T, Lee MG, and Hayashi Y. (2017). Hippocampus-dependent goal localization by head-fixed mice in virtual reality. eNeuro 4, e0369–16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheintuch L, Rubin A, Brande-Eilat N, Geva N, Sadeh N, Pinchasof O, and Ziv Y. (2017). Tracking the same neurons across multiple days in Ca2+ imaging data. Cell Rep. 21, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD (2015). Spatial navigation in autism spectrum disorders: a critical review. Front. Psychol 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa T, Asai H, Ohkawa N, Nomoto M, Okubo-Suzuki R, Ghandour K, Sato M, Hayashi Y, Inokuchi K, and Fukai T. (2017). Automatic sorting system for large calcium imaging data. bioRxiv. 10.1101/215145. [DOI] [Google Scholar]
- Tsao A, Moser MB, and Moser EI (2013). Traces of experience in the lateral entorhinal cortex. Curr. Biol 23, 399–405. [DOI] [PubMed] [Google Scholar]
- Valero M, Cid E, Averkin RG, Aguilar J, Sanchez-Aguilera A, Viney TJ, Gomez-Dominguez D, Bellistri E, and de la Prida LM (2015). Determinants of different deep and superficial CA1 pyramidal cell dynamics during sharpwave ripples. Nat. Neurosci 18, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ham IJM, Martens MAG, Claessen MHG, and van den Berg E. (2017). Landmark agnosia: evaluating the definition of landmark-based navigation impairment. Arch. Clin. Neuropsychol 32, 472–482. [DOI] [PubMed] [Google Scholar]
- Vogelstein JT, Packer AM, Machado TA, Sippy T, Babadi B, Yuste R, and Paninski L. (2010). Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol 104, 3691–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen X, Lee H, Deshmukh SS, Yoganarasimha D, Savelli F, and Knierim JJ (2018). Egocentric coding of external items in the lateral entorhinal cortex. Science 362, 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener SI, Paul CA, and Eichenbaum H. (1989). Spatial and behavioral correlates of hippocampal neuronal activity. J. Neurosci 9, 2737–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, and McNaughton BL (1993). Dynamics of the hippocampal ensemble code for space. Science 261, 1055–1058. [DOI] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, et al. (2012). Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486, 261–265. [DOI] [PubMed] [Google Scholar]
- Youngstrom IA, and Strowbridge BW (2012). Visual landmarks facilitate rodent spatial navigation in virtual reality environments. Learn. Mem 19, 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba JD, Diamantopoulou A, Danielson NB, Grosmark AD, Kaifosh PW, Bowler JC, Liao Z, Sparks FT, Gogos JA, and Losonczy A. (2017). Impaired hippocampal place cell dynamics in a mouse model of the 22q11.2 deletion. Nat. Neurosci 20, 1612–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, and Schnitzer MJ (2013). Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci 16, 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and custom computer code associated with this study will be made available by the authors upon request.