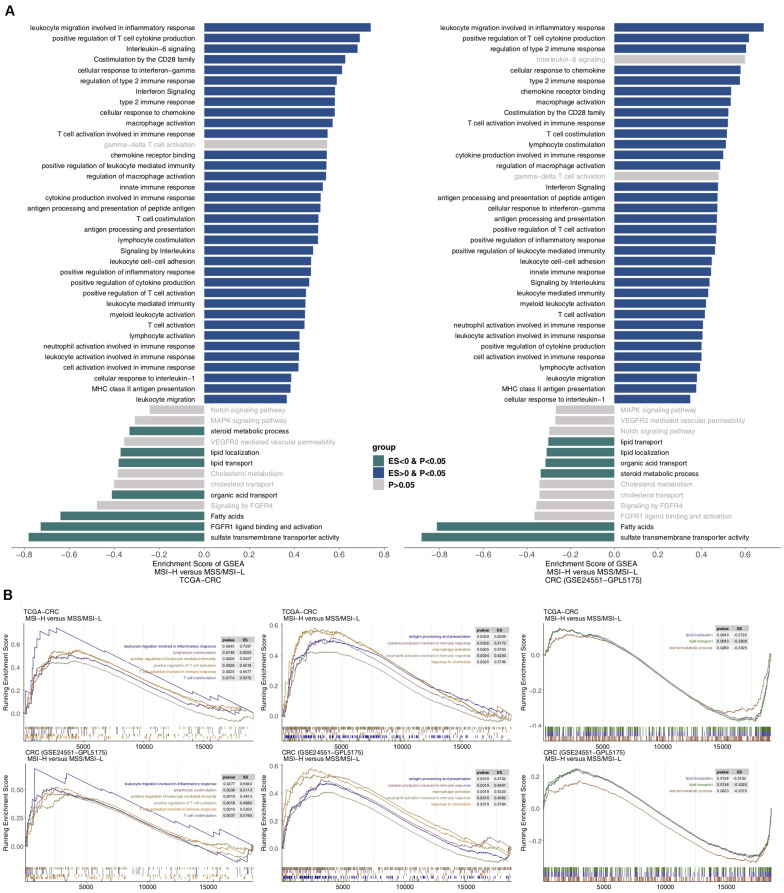
FIGURE 5.
Transcriptomic analysis of the biological function traits of MSI-H and MSS/MSI-L tumors in the TCGA-CRC cohort and another CRC cohort (GSE24551). (A) Differences in pathway activities scored by GSEA between MSI-H and MSS/MSI-L tumors in the TCGA-CRC cohort. Enrichment results with significant differences between MSI-H and MSS/MSI-L tumors are shown. A blue bar indicates that the ES of the pathway is more than 0, while a green bar indicates that the ES of the pathway is less than 0. (B) GSEA of hallmark gene sets downloaded from the MSigDB. All transcripts are ranked by the log2 (fold change) between MSI-H and MSS/MSI-L tumors in the TCGA-CRC cohort and another CRC cohort (GSE24551). Each run was performed with 1,000 permutations. Enrichment results with significant differences between MSI-H and MSS/MSI-L tumors are shown. GSEA, gene set enrichment analysis; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; TMB, tumor mutational burden; MSS/MSI-L, microsatellite-stable/microsatellite instability-low; MSI-H, microsatellite instability-high; ES, enrichment score; and MSigDB, Molecular Signatures Database.