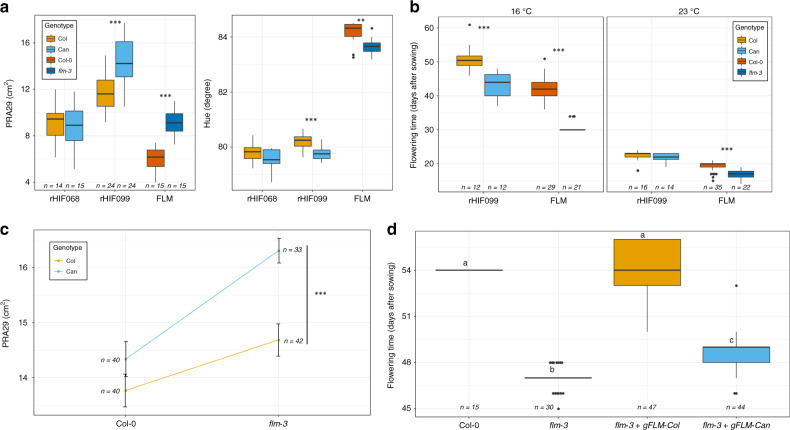
Fig. 1. Natural variation at FLM is responsible for the QTLs observed at the end of chromosome 1.
a Boxplots comparing PRA29 or Hue in alternate fixed alleles at the segregating region of the two most informative recombinant lines (rHIF068 is fixed at FLM but segregates elsewhere, and rHIF099 is segregating at FLM, see their exact genotype in Supplementary Data 4), and in flm-3 versus its wild-type background Col-0. b Modulation of flowering time according to the growth conditions at 16 and 23 °C (in long days) in rHIF099 lines and flm-3 versus Col-0. c Quantitative complementation assay assessing the effect on PRA29 of each of the possible genotypic combination between the Can or Col alleles at the QTL (in the rHIF background) and the wild-type or mutant (flm-3) alleles at FLM (in the Col-0 background). PRA29 was obtained using the Phenoscope. Error bars indicate the standard error of the mean. Significance is shown for the effect of the QTL allele in complementing flm-3. d Functional complementation based on flowering time of the flm-3 mutant transformed with the genomic fragments of FLM (gFLM) from Col-0 or Can-0. Source data are provided as a Source Data file.