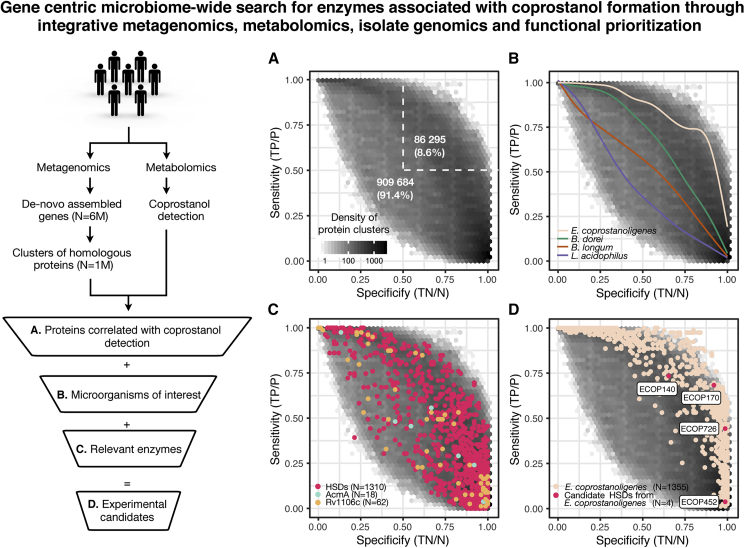
Figure 2.
Integrative Analysis of Metagenomes, Metabolomes, Isolate Genomes, and Enzymatic Functions Reveals Candidate Bacterial Genes Involved in Cholesterol Metabolism in the Human Gut Microbiome
Human gut microbiome genes from a de novo assembled gene catalog, after additional clustering step into groups of homologous proteins (at least 50% aa identity), were correlated with coprostanol detection in paired metagenomic-metabolomic samples and further prioritized by incorporating information from relevant microorganisms and enzymes.
(A) Scores of specificity and sensitivity in relation to presence of coprostanol were calculated for each cluster of homologous proteins, and their density is represented through hexagonal bin plot; 8.6% of protein clusters are found with greater than 50% specificity and sensitivity to coprostanol detection.
(B) Proteins encoded by gut microbes of interest (implicated in coprostanol formation in the literature) were used to query the clusters of homologous proteins. Clusters containing proteins with >50% aa identity to proteins found within a specified organism were used to generate a smoothed trend line (see Figure S1E for the location of species matched clusters). According to the location of these trend lines, E. coprostanoligenes matching clusters are more specifically associated with coprostanol formation than clusters from other microbes.
(C) Clusters of homologous proteins were queried with characterized enzymes known to either catalyze the oxidation of cholesterol to cholestenone: cholesterol oxidases (PF09129), AcmA from S. denitrificans, and Rv1106c from M. tuberculosis or enzymes that can perform very similar chemical transformations (HSDs: RUMGNA_00694, Elen_1325, Elen_0198, and KGH18088). USEARCH ublast (Edgar, 2010) analysis was performed with inclusive cutoffs (>25% aa identity and 50% coverage).
(D) Combining the evidence from (A)–(C), 4 putative HSDs in E. coprostanoligenes were identified, 3 of which (ECOP170, ECOP726, and ECOP442) had high specificity with regard to the presence of coprostanol (>0.9), albeit with greatly varying sensitivity. All four enzymes were chosen for further biochemical validation.
All panels based on dataset 1 analysis; see Figures S1A–S1D for dataset 2 analysis. See also Figure S1 and Table S1.