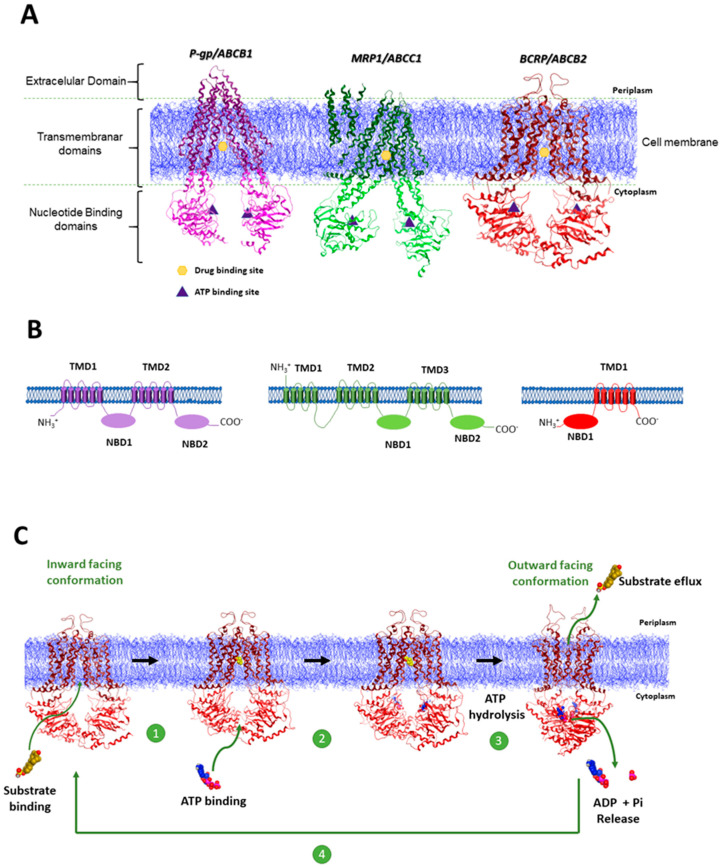
Figure 1.
(A) High resolution 3D structures of the mouse P-gp (PDB-ID:5KPI) [20], which has 87% sequence identity to human P-gp; bovine MRP1(PDB-ID:5UJ9) [21], which has 91% protein identity with the human protein; and human BCRP (PDB-ID:5NJ3) [22]. The NBD domains for each protein are depicted in light purple (P-gp), light green (MRP1) and light red (BCRP), whereas the TMD domains are depicted in the dark variations of the same colors. Cell membrane is depicted in blue. Adapted from: Nat Rev Cancer 18, 452–464 (2018) [9]; (B) Membrane topology models of P-gp, MRP1, and BCRP proteins. P-gp possesses two homologous halves, each containing a TMD consisting of six putative membrane-spanning α-helices at the N-terminus and a cytosolic hydrophilic NBD at the c-terminus that are involved in binding and hydrolyzing ATP. MRP1, in addition to the two NBD and two TMD containing six α-helices, is composed by and extra TMD consisting of five α-helices. The half transporter BCRP contains one TMD of six α-helices and one NBD. The ATP-binding site of this transporter is found on the amino-terminal side (N) in contrast to P-gp and MRP1. Was adopted the same color scheme used in A. (C) Simplified representation of the ABC drug efflux mechanism using the 3D structure of BCRP: 1—Substrate (molecule in yellow) enters in the drug binding site localized within the TMD domains of the transporter and triggers ATP (molecules in blue and pink) binding; 2—ATP binds to the NBD of the transporter; 3—Following ATP binding is induced the formation of the NBD sandwich dimer. ATP hydrolysis triggers conformational changes resulting in an outward open TMD conformation with reduced affinity to the substrate allowing the drug extrusion. 4—ADP + Pi release induces the resetting of the transporter to its basal inward facing conformation. To build this picture were used Cryo-EM structures of BCRP: structure of BCRP with the substrate (estrone-3-sulfate) placed in the DBS (PDB-ID:6HCO) [23]; 2 ATP molecules were placed in the NBD using MOE.; structure of BCRP in the outward facing configuration and two ATP molecules placed in the NBD (PDB-ID:6HBU) [23].