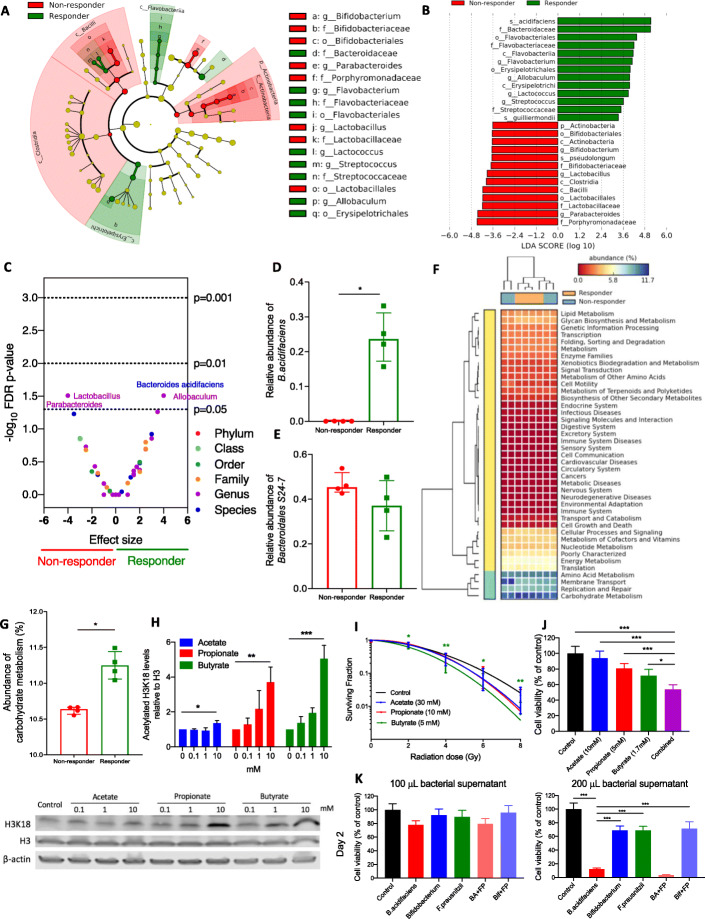
Fig. 5.
Differences in composition of the gut microbiome between responders and non-responders. a Taxonomic cladogram from LEfSe showing differences among taxa between responders and non-responders in the soluble HF group. Dot size is proportional to the abundance of the taxon. b Linear discriminant analysis (LDA) scores computed for differentially abundant taxa in the microbiomes of responders (green) and non-responders (red). Length indicates the effect size associated with a taxon, p = 0.05 for the Kruskal-Wallis test. c Discrete false-discovery rate of different abundant taxa in responders and non-responders in the soluble HF group. Differential abundance within all taxonomic levels in responders versus non-responders by Mann-Whitney U test. Dots are overlapping between Bacteroides acidifaciens and Allobaculum, and between Lactobacillus and Parabacteroides. Relative abundance of d B. acidifaciens and e Bacteroidales S24-7 and in responders and non-responders in the soluble HF group. f, g Metagenomic functional prediction by PICRUSt of the gut microbiome in responders (n = 4) and non-responders (n = 4) in the soluble HF group with reference to the KEGG database level 2. Columns represent mice (responders, orange; non-responders, blue), and rows represent enrichment of predicted KEGG pathways (red, low enrichment; yellow, medium enrichment; blue, high enrichment). h Western blot analysis of histone acetylation levels of RT112 cells treated with SCFAs (N = 3). i Linear quadratic survival curves of IC10-treated RT112 cells with receiving irradiation of 0–8 Gy (N = 3). j Cell survival analysis of RT112 cells treated with single SCFA and combined SCFAs mixture (N = 3). Combined (purple bar) denotes SCFA mixture of 10 mM acetate, 5 mM propionate, and 1.7 mM butyrate. k Reduced cell survival of RT112 cells by bacterial supernatants at day 2 (N = 1). BA+FP denotes the cross-feeding of B. acidifaciens and F. prausnitzii, while Bif+FP denotes the cross-feeding of Bifidobacterium and F. prausnitzii. *p < 0.05; **p < 0.01; ***p < 0.001