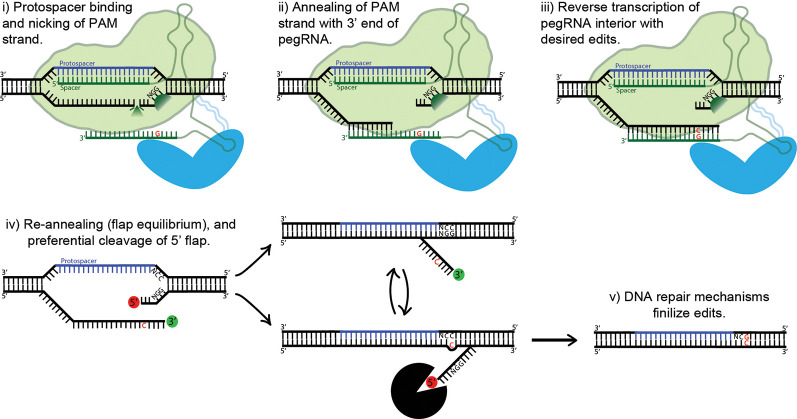
Figure 5.
Proposed mechanism of prime editing. First, the 5’ end of the pegRNA binds to the protospacer of the target DNA and the protospacer-adjacent motif (PAM) strand is nicked (i). The nicked PAM strand then hybridizes with the primer binding site (PBS) at the far 3’ end of the pegRNA (ii). The interior of the pegRNA then serves as a template for reverse transcription, which extends from the free 3’-OH of the PAM strand (iii). The prime editing complex then disengages, leaving the target site with two redundant PAM strands, or “flaps” (iv). The unedited 5’ flap is preferentially degraded by cellular endonucleases, allowing the edited 3’ flap to hybridize with the non-PAM strand. Finally, DNA repair mechanisms transfer the desired edits to the non-PAM strand (v).