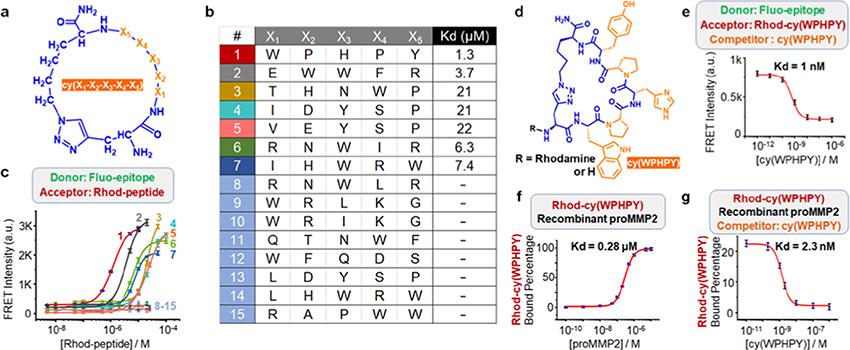
Figure 2.
Validation of the hit sequences generated from library screening. (a) Generic structure of the cyclic peptide library. (b) Identified hits from the screening. (c) Förster resonance energy transfer (FRET) signals generated from the binding between 100 nM fluorescein-labeled epitope (Fluo-epitope) and varying concentrations of rhodamine B-modified ligand (Rhod-peptide). (d) Structure of cy(WPHPY) with and without the rhodamine B tag. (e) Competitive FRET results obtained using 100 nM Fluo-epitope, 100 nM Rhod-cy(WPHPY), and varying concentrations of cy(WPHPY) as the donor, acceptor, and competitor, respectively. The interaction between unlabeled cy(WPHPY) and Fluo-epitope displaced Rhod-cy(WPHPY), causing decreased FRET signals. (f) Fluorescence polarization (FP) assay results demonstrating the binding affinity between Rhod-cy(WPHPY) (100 nM) and the full-length recombinant proMMP2. (g) Competitive FP results demonstrating the binding between the unlabeled cy(WPHPY) and recombinant proMMP2 (100 nM). Here, the unlabeled cy(WPHPY) displaced Rhod-cy(WPHPY) (100 nM) and led to decreased FP values.