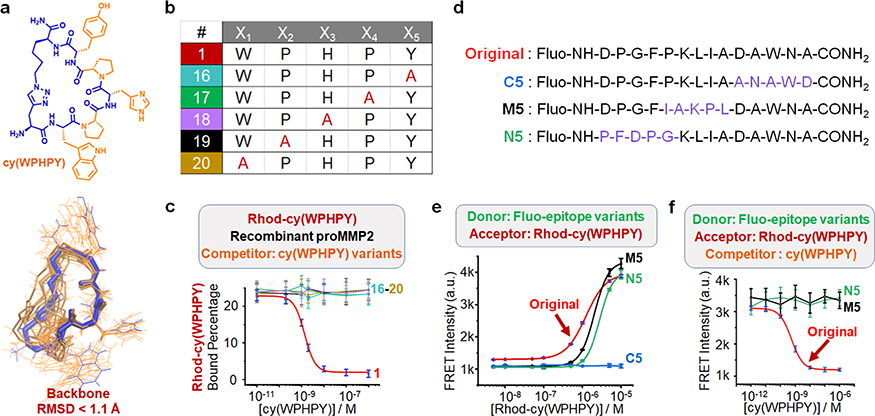
Figure 3.
Analysis of the structure–activity relationship. (a) Optimal conformations of cy(WPHPY) as calculated using molecular dynamics (MD) approaches. The blue wire is the most representative conformation generated from the conformation search. The orange wires illustrate the conformations that were evenly extracted every 2 ns from the MD simulation trajectory. (b) Alanine scanning variations of cy(WPHPY). (c) Competitive FP results obtained using alanine-substituted cy(WPHPY) variations. The competitive FP results of cy(WPHPY) were included as a reference (red line, peptide 1). (d) Partially scrambled epitope sequences in comparison with the original epitope. (e) FRET signals generated from the binding between 100 nM Fluo-epitope variations and varying concentrations of Rhod-cy(WPHPY). The result from the original Fluo-epitope was included as a reference. C5 was not able to bind to Rhod-cy(WPHPY), while M5 and N5 retained the binding affinity. (f) Competitive FRET assay results demonstrating the lack of binding affinity between the unlabeled cy(WPHPY) and the partially scrambled epitopes (M5 and N5). Unlabeled cy(WPHPY) could not displace Rhod-cy(WPHPY) from the epitope; therefore, the FRET intensities did not change. The result from the original Fluo-epitope was included as a reference.