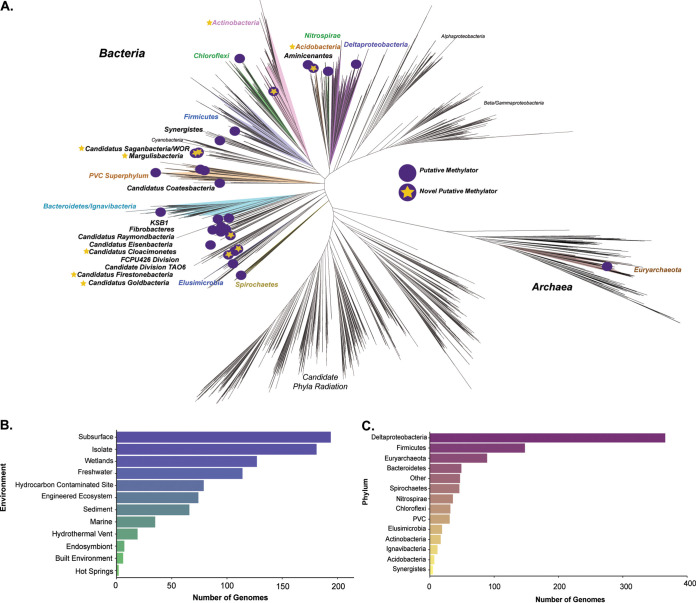
FIG 1.
Phylogenetic distribution and diversity of putative methylators. (A) Putative methylators among the prokaryotic tree of life. The highest quality putative methylator from each identified phylum was selected as a representative among select references from Anantharaman et al. (39) from across the tree of life. The tree was constructed from an alignment of 16 concatenated ribosomal proteins. Purple dots represent putative methylating phyla, and those with stars represent phyla containing putative novel methylating lineages that, to our knowledge, have not been identified as potential mercury methylators before this study. Groups in bold and/or colored are putative methylating groups, whereas a few uncolored groups in nonboldface font are noted only for orientation. (B) Numbers of putative methylating genomes found in various environments or genomes sequenced from isolates. (C) Numbers of putative methylating genomes belonging to each phylum. Phylum names are given for all groups except the Deltaproteobacteria class of Proteobacteria, as no putative Proteobacteria methylators were identified outside the Deltaproteobacteria. The group “other” contains several candidate phyla and genomes within groups with few representatives, described in Fig. S1 in the supplemental material.