Fig. 1.
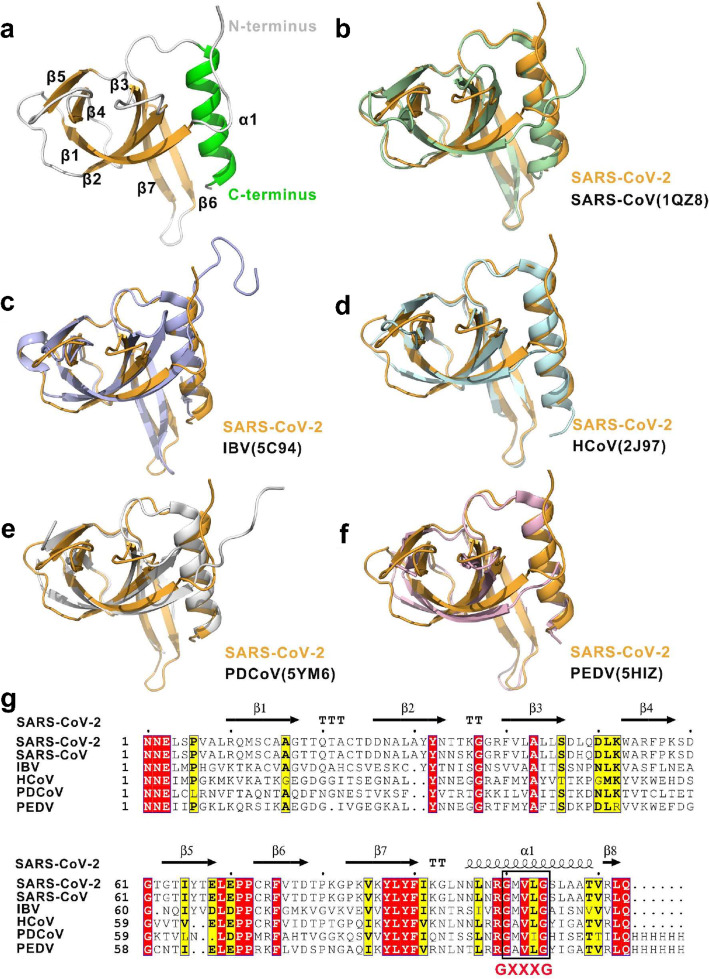
Overall structure of the SARS-CoV-2 nsp9 protomer. a Different views of the protomeric architecture of SARS-CoV-2 nsp9 with the secondary structure labeled. The α-helix, β-sheets, and loops are colored in green, orange, and white, respectively. b–f Superimposition of SARS-CoV-2 nsp9 with other nsp9 structures in the coronavirus family. The nsp9s from SARS-CoV-2, SARS-CoV, avian infectious bronchitis virus (IBV), human coronavirus 229E (hCoV-229E), porcine delta coronavirus (PDCoV), and porcine epidemic diarrhea virus (PDEV) are colored in orange, light blue, marine, white, and salmon, respectively. The PDB codes are indicated in the lower right corner. gSequence alignment of CoV nsp9 homologs. Comparison of the SARS-CoV, IBV, HCoV, PDCoV, and PDEV sequences with the SARS-CoV-2 sequence. Identical residues are highlighted in red, and conserved residues are shown in yellow. The table was produced with ESPript 3.0, using secondary structure elements for SARS-CoV-2 nsp9 assigned using DSSP. Residues boxed in red are completely conserved