Fig. 3.
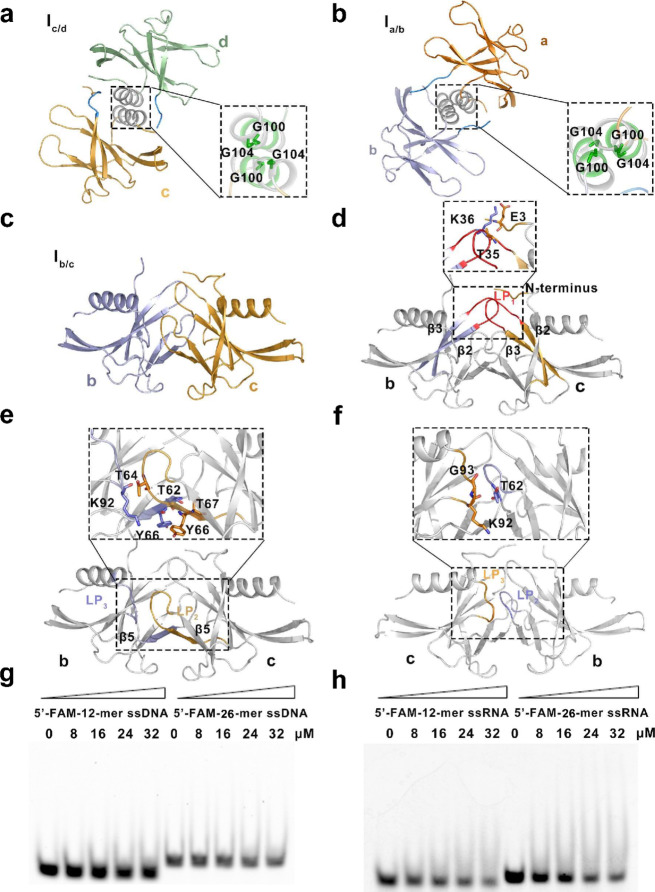
Details of structure of the SARS-CoV-2 nsp9 tetramer and function in nucleic acid binding. a, b Details of Ic/d and Ia/b in the helix interface. The molecules in these two interfaces are shown as cartoons and colored and labeled as in Fig. 2a. The expanded boxed area to the right shows a detailed view of the critical residues in the Ic/d and Ia/b interfaces, which are shown as sticks. c Molecules involved in Ib/c. The molecules in this interface are shown as cartoons and colored and labeled as in Fig. 2a. d–f Detailed views of the first, second, and third contact regions in interface Ib/c, respectively. The secondary structures of each contact region are colored and labeled according to Fig. 2a and the critical residues in each region are shown as sticks. g Single-stranded DNA (ssDNA)-binding abilities of SARS-CoV-2 nsp9, as determined by electrophoretic mobility shift assay. h Single-stranded RNA (ssRNA)-binding abilities of SARS-CoV-2 nsp9. The length of ssDNA, the state of SARS-CoV-2 nsp9, and the concentration of proteins used in the reaction system are indicated above the gel