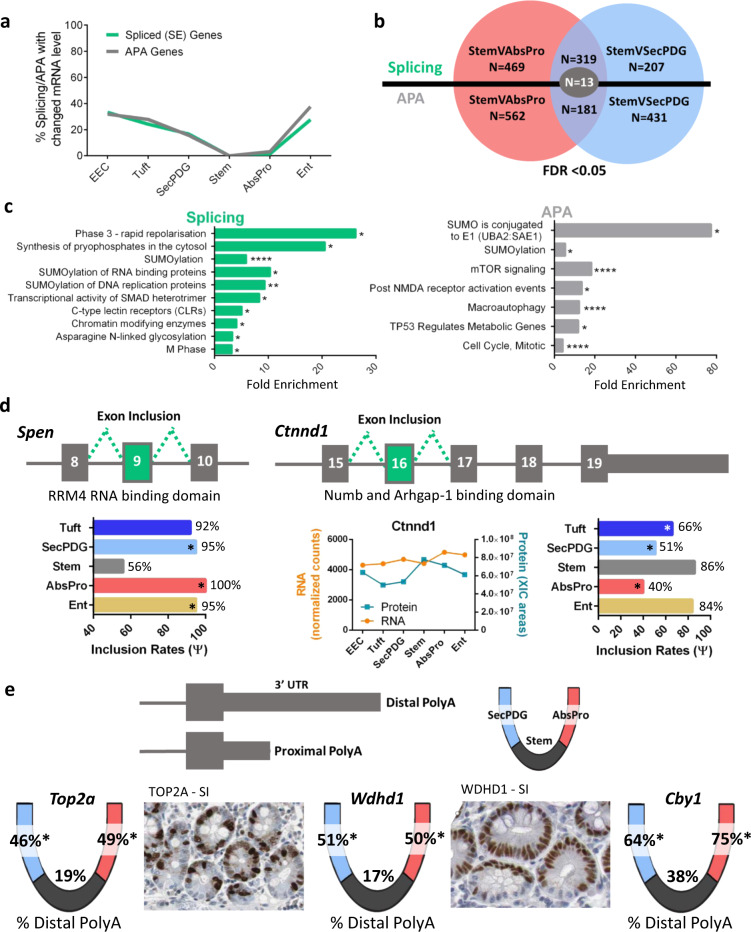
Fig. 4. Alternative splicing and polyadenylation changes that occur during intestinal crypt loss of stemness.
a Percentage of alternatively spliced (FDR < 0.05) or polyadenylated genes (padj < 0.05) that also change gene expression (padj < 0.01) compared with stem. b Venn diagram overlap of APA and alternatively spliced genes between stem versus AbsPro and stem versus SecPDG (FDR < 0.05). n = 13 genes were both APA and alternatively spliced differently in stem cells versus SecPDG and stem cells versus AbsPro (Supplementary Fig. 21a). c Gene ontology (reactome pathway) analysis of the commonly spliced genes when comparing stem versus AbsPro and stem versus SecPDG (rMATS; n = 332 (319 + 13) genes) and common APA genes (DaPars, n = 194 (181 + 13) genes); FDR < 0.05. Sumoylation and cell cycle ontologies of alternatively processed genes are common to both splicing and APA changes (Supplementary Fig. 21b, c). d Two examples of alternatively spliced genes, Spen and Ctnnd1, which are differentially processed in stem cells versus progenitor cells. The exon inclusion rate for each event is shown in the bar graph. Ctnnd1 protein was detected by MS and the abundance compared with mRNA is displayed. e Three examples of genes with significant changes in alternative polyadenylation choice: Top2a, Wdhd1, and Cyb1. Overlaid on the crypt-base diagram are the percentage of distal polyA usage for each of the genes in the three cryptal cell compartments: stem, AbsPro, and SecPDG. Human protein atlas images show strong small intestine (SI) staining patterns of TOP2A and WDHD1 in the transit amplifying zone but a lack of staining in the stem cell niche despite the fact that Top2a mRNA levels are elevated in stem cells compared to progenitor cells and Wdhd1 mRNA levels are the same among the cell types (Supplementary Fig. 22). Additional immunohistochemistry images of human intestine are provided in Supplementary Fig. 23. FDR significance is defined by: *<0.05, **<0.01, ***<0.005, ****<0.001.