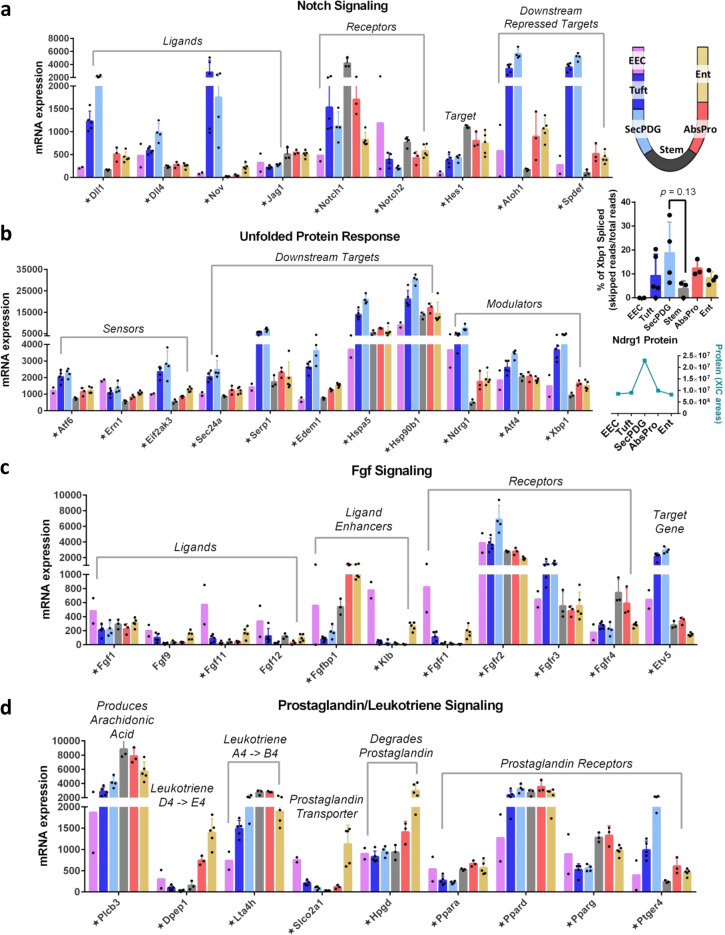
Fig. 6. Lineage commitment to secretory and absorptive lineages are influenced by signaling pathways.
a mRNA expression of Notch ligands (elevated in SecPDG/tuft), receptors (elevated in stem), and downstream targets. b mRNA expression of unfolded protein response (UPR) components including sensors, modulators, and downstream targets are elevated in SecPDG. Xbp1 activation, determined by a cytoplasmic splicing event, is elevated in SecPDG (inset—unpaired two-sided t test). Modulator Ndrg1 was detected via MS and shows protein is elevated in SecPDG (inset graph) consistent with mRNA expression (Ndrg1 protein was not detected in Stem). c mRNA expression of Fgf signaling components including ligands (showing some EEC expression), ligand enhancer Fgfbp1 expressed in AbsPro and Ent, receptors (well expressed in all cells, highest in SecPDG and tuft) and target gene (highly expressed in SecPDG and tuft). d Prostaglandin and leukotriene precursors and final products are produced by tuft cells (Supplementary Fig. 25), but absorptive lineage cells AbsPro and Ent, might contribute to the production (Plcb3, Dpep1, and Lta4h) and degradation (Slco2a1 and Hpgd) of prostaglandin signals. Prostaglandin receptor Ptger4 is enriched in SecPDG, whereas alternate receptors Pparg and Ppara are enriched in the absorptive lineage. Star annotation by gene name symbolizes significant differential mRNA expression in at least one cell type compared with stem (padj < 0.01). mRNA expression values are normalized counts and error bars are standard deviation. mRNA differential expression analysis was performed with the following biological replicate numbers: stem = 3, AbsPro = 3, SecPDG = 4, tuft = 5, Ent = 5, and EEC = 2.