Fig. 5. Mechanism of SR.
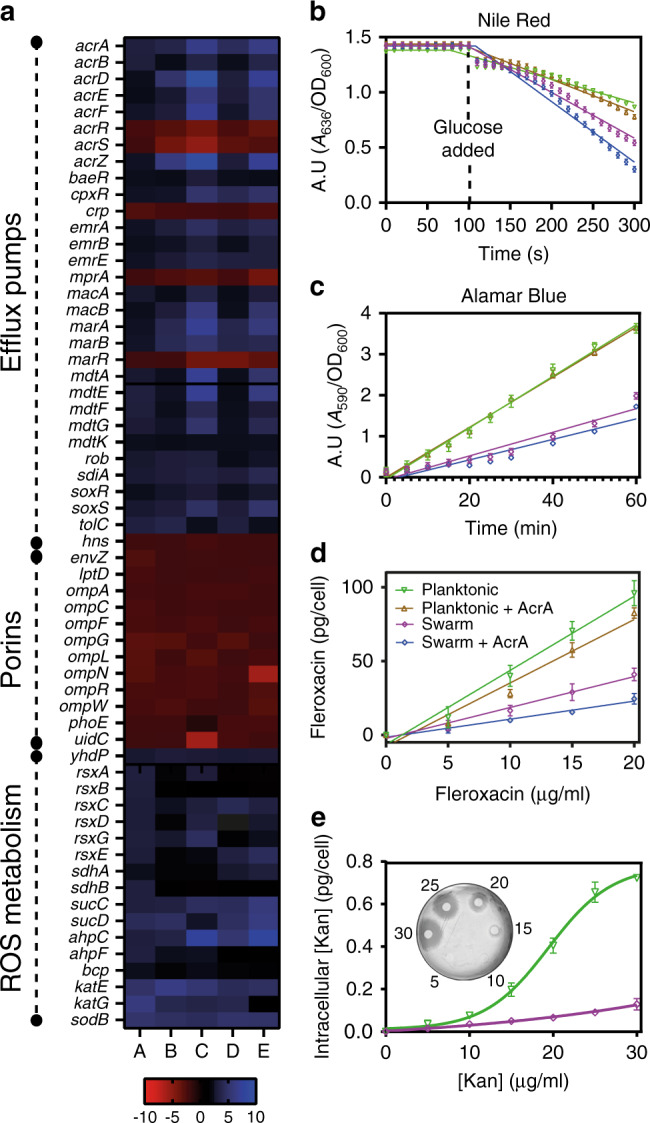
a Comparison of log2 fold changes in gene expression. (A) Swarm vs. Planktonic, (B) Swarm + Kan20 vs. Swarm, (C) Swarm + Kan20 + AcrA vs. Swarm, (D) Swarm + Cip2.5 vs. Swarm, (E) Swarm + Cip2.5 + AcrA vs. Swarm. The concentration of AcrA was 0.1 μg/ml, and the log2 fold change cutoff value was 2. About 25 genes encoding efflux pump components representing all five classes of pumps and their regulators were upregulated in swarm cells (~2–3 fold), while ten genes expressing different porins or their regulators were downregulated (~2–3 fold). Except for porins, all these genes were further upregulated (~2–6 fold) in swarm cells under antibiotic stress (Kan and Cip). The presence of AcrA also increased expression of these same genes (~2–9 fold). Twenty-three genes for energy metabolism (~2–4 fold) and 16 genes related to ROS catabolism (~2–6 fold) were also upregulated. b Efflux assay using Nile red indicator dye (n = 3). Glucose was added at 100 s. Nonlinear regression analysis of Planktonic, Planktonic + AcrA, Swarm, and Swarm + AcrA data sets (see d for color key) yielded rate constants of 1.001 × 10−0.6, 8.669 × 10−0.7, 6.630 × 10−0.6, and 1.165 × 10−0.6, respectively. A.U. = (A636/OD600). The time required for 50% Nile Red efflux or teff50, was significantly low in swarm cells (230 s for swarm and ≥300 s for planktonic cells; compare purple vs. green lines; p < 0.0001 for the two values). Addition of AcrA yielded tefflux50 with a 1/10th increase in swarm cells (207 s, p = 0.0003 for Swarm + AcrA vs. Swarm from a two-tailed T test) and 1/20th increase in planktonic cells (282 s, compare brown vs. blue lines; p = 0.0038 for planktonic +AcrA vs. Planktonic from a two-tailed T test). c Membrane permeability assay using Alamar Blue indicator dye (n = 3). Nonlinear regression analysis of Swarm, Swarm + AcrA, Swim, and Swim+ AcrA data sets (see d for color key) produced slopes of 0.06254, 0.06103, 0.02876, and 0.02498 respectively. A.U. = (A590/OD600). d Estimated intracellular concentration of Fleroxacin (n = 3). The slopes of the fitted lines of data sets for Swarm, Swarm + AcrA, Planktonic, and Planktonic + AcrA were 2.090, 1.220, 5.010, and 4.290, respectively. e Estimation of intracellular antibiotic concentration in E. coli using the Disc-diffusion assay. See “Methods” for details. The inset image shows one representative plate with discs containing planktonic samples incubated with the Kan concentrations indicated by the numbers. Inhibition zones around the filter discs were compared to those from a standard curve generated with known concentrations of kanamycin. Swarm cells show ~8-fold decrease in intracellular [Kan] at Kan30 compared to planktonic cells. Data are presented as mean values ± SD in (b–e). Source data are provided as a Source Data file.
