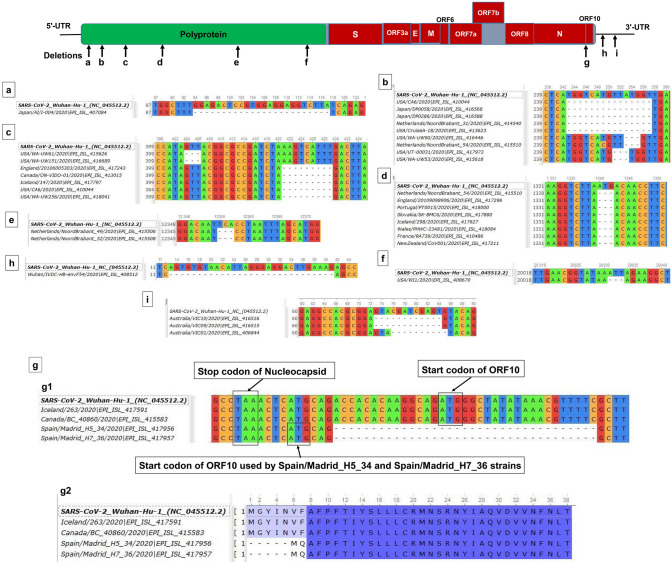
Figure 1.
Genomic deletion analysis of SARS-CoV-2. Genomic deletion analysis of SARS-CoV-2 strains identified (a) 24 (nt) deletions in NSP1 in a Japanese strain; (b) 15-nt deletions in NSP1 of viral strains from USA, Japan and the Netherlands alongside three-nt deletions in USA and Netherlands; (c) three-nt deletion in NSP1 of American strains and very adjacent to that, nine-nucleotide deletion of strains from the USA, England and Canada and Iceland; (d) three-nucleotide deletions in NSP2 were observed in 99 strains from Netherlands, England, Portugal, Slovakia, Iceland, Wales, France and New Zealand (representatives from each countries were shown); (e) NSP8 undergoes three-nt deletion in Netherlandian/dutch/hollanders strains; (f) three-nucleotide deletion in NSP15 of USA strain; (g-g1) 35nt deletion, including start codon position of ORF10 of Spain strains, and the start codon in spacer position, has been used for ORF10 coding; and as a result, (g-g2) five aa residues deletion in those strains starting from position 1 to 5. Deletion of (h) 29-nt reported from Wuhan, and (i) 10-nt in 3′-UTR of strains belonged to Australia. The position of nt represents the starting position from each ORFs, for instance, position of ORF1ab was considered for the NSPs. MAFFT online tool36 was used for alignment, and Unipro-UGENE40 used for visualization.