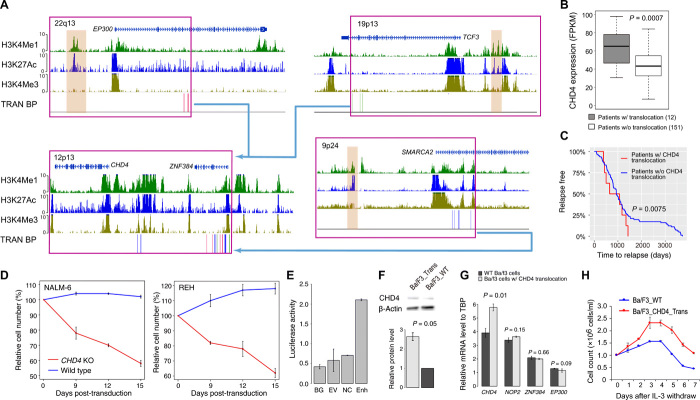
Fig. 2. Enhancer hijacking to CHD4 by translocation in patients with B-ALL.
(A) Translocations result in enhancer hijacking to CHD4 in B-ALL. Three different translocation partners are identified in the cohort. Histone modification ChIP-seq data of human CD19+ B cells (identifying enhancers), SNVs, and SV break points (BPs; each vertical line represents a patient) are shown. The hijacked enhancers are highlighted in brown. (B) Expression levels of CHD4 in patients with and without translocations. P value of one-sided t test is shown (n = 163). (C) Patients with CHD4 translocation have shorter time to relapse. P value of log-rank test is shown (n = 163). (D) CHD4 knockout impairs growth of B-ALL cell lines, NAML-6 and REH. (E) Luciferase reporter assay of hijacked enhancer. BG, no DNA vector control; EV, vector containing no enhancer; NC, negative control; Enh, test EP300 enhancer. (F) Western blots of CHD4 protein in Ba/F3 cells with and without translocation. P value of one-sided t test is shown (n = 2). (G) mRNA levels of CHD4, NOP2, ZNF384, and EP300 in Ba/F3 cells with and without translocations. P values were calculated using one-sided t test (n = 2 for all tests). TBP, TATA-box Binding Protein. (H) Ba/F3 cells with induced translocation undergo oncogenic transformation. Enhancer hijacking translocation was induced using CRISPR-Cas9. Error bars represent SD.