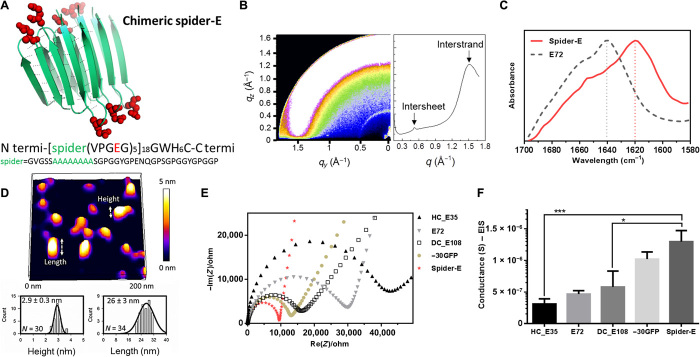
Fig. 3. Sequence, structure, and proton conduction of recombinant supercharged spider-E thin films on IDE.
(A) Rationally designed supercharged spider silk–inspired proteins (spider-E). The spider motif contains a poly-A sequence (green) and anionic supercharged regions (red) that are forming the loops between the rigid β sheets. (B) Structure analyses of the spider-E supported film. Two peaks were detected by GIXD, indicating the characteristic intersheet and interstrand distances, respectively. (C) FTIR characterization of the films indicate random coils for the E72 sample (gray dashed line) with an amide I peak located at 1640 cm−1 and a shift to a typical β sheet amide I peak for the spider-E sample (red solid line) at 1620 cm−1. (D) Morphology analyses of the spider-E supported film. Quantification of the nanostructures assembled through spider β sheet domains by AFM. This sample was obtained by extensive swelling of the film by water contact to induce separation between the domains. (E) Nyquist plots obtained at RH = 90% for the five genetically engineered samples including spider-E. The impedance curve of the spider-E sample shows the lower resistance value among all the samples (red). (F) Comparison of conductance of the resulting devices demonstrating the stepwise increase in the transport properties due to the improved protein design. The proton transport of spider-E thin films on IDEs is noticeably higher than HC_E35 (***P = 0.0009, n > 3) and DC_E108 (*P = 0.0155, n > 3).