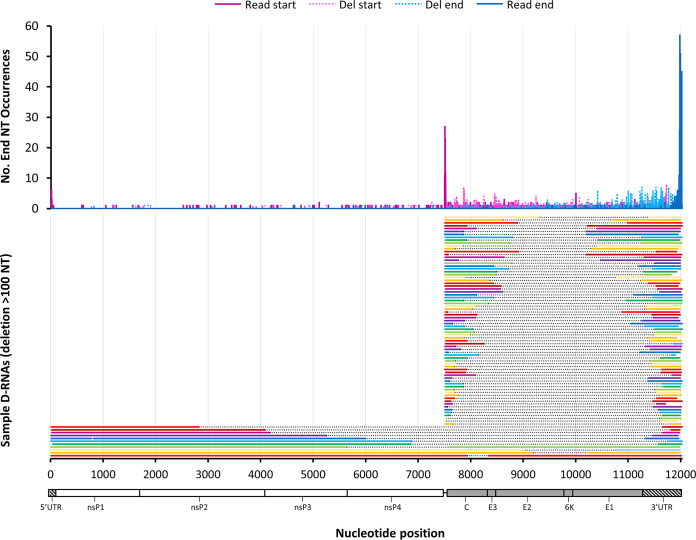
FIG 5.
Oxford Nanopore’s direct RNA sequencing of intracellular CHIKV D-RNA species. Intracellular RNA from CHIKV-infected Vero cells was sequenced using Oxford Nanopore’s direct RNA sequencing kit on a MinION sequencer, reads were mapped using minimap2, and then read boundaries for CHIKV-mapped reads were extracted from SAM files. Boundary counts for defective RNA reads containing deletions were enumerated and plotted (top), and the raw single reads containing the largest deletions for genomic and subgenomic D-RNAs are shown (bottom).