Fig. 3. Synthetic attC sites embedded into protein coding regions.
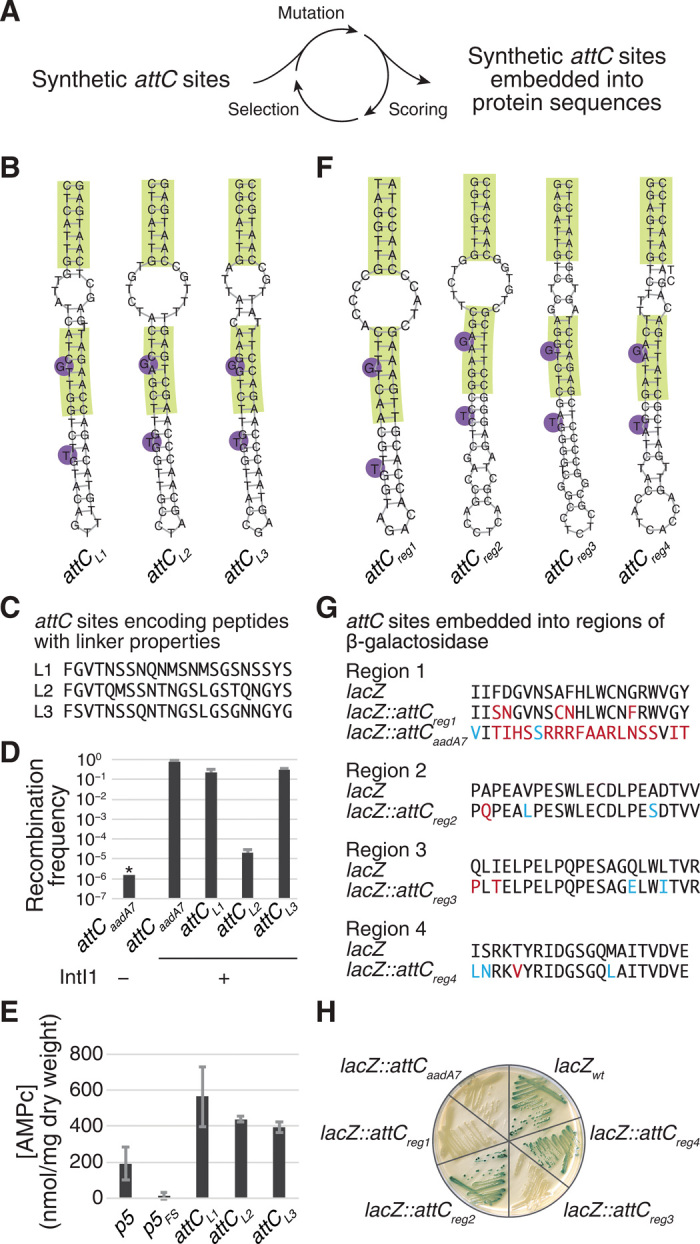
(A) In silico directed evolution approach that was used to generate synthetic attC sites with peptide linker properties (B-E) and to embed synthetic attC sites into lacZ (F-H). (B) Structures of the three synthetic attC sites encoding peptide linkers L1, L2, and L3, predicted using ViennaRNA 2.1.8. (C) Protein sequences of encoded peptide linkers. (D) Recombination frequencies of attCaadA7 (positive control) and synthetic attC sites encoding peptide linkers. Values represent means of three independent experiments; error bars represent mean absolute error. Asterisk (*) indicates that the recombination frequency was below detection level, indicated by the bar height. (E) Results of a bacterial two-hybrid assay with the two domains of Bordetella pertussis adenylate cyclase fused either with a natural linker (p5, positive control), a natural linker with a frameshift mutation (p5FS, negative control), or synthetic attC sites encoding peptide linkers. Values represent means of three independent experiments; error bars represent mean absolute error. (F) Predicted structures of the synthetic attC sites embedded into four regions of the lacZ gene encoding β-galactosidase, predicted using ViennaRNA 2.1.8. (G) Protein sequences of the four β-galactosidase target regions and sequences after attC site embedding. Blue, mutations that preserve the amino acid physicochemical properties; red, other nonsilent mutations. (H) Strains with the four synthetic attC sites embedded into lacZ and streaked on an LB agarose plate with X-gal and isopropyl-β-D-thiogalactopyranoside. Blue color indicates a functional β-galactosidase, as in lacZwt. White color indicates that an embedded attC site perturbed the function of the β-galactosidase, as in lacZ::attCaadA7.
