Fig. 1 |. In vivo neuroepigenome-editing tools.
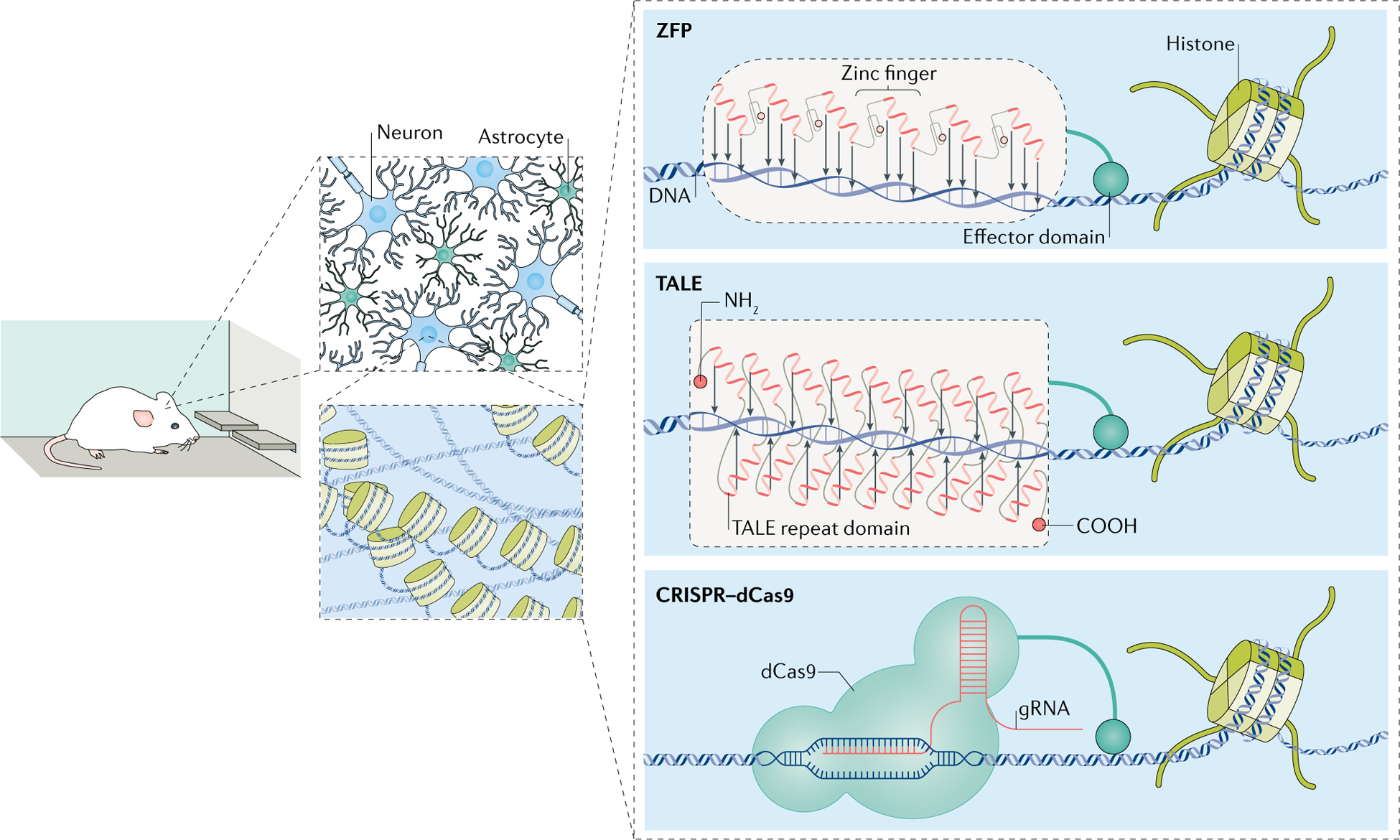
Zinc-finger proteins (ZFPs) and transcriptional activator-like effectors (TALEs) are DNA-binding proteins that can be engineered to target specific genomic loci. Fusing the DNA-binding domain to transcription effector proteins offers a tool to selectively regulate gene transcription. In the clustered regularly interspaced short palindromic repeats (CRISPR) system, a nuclease-dead CRISPR-associated protein 9 (dCas9) molecule fused to an effector domain forms a complex with a guide RNA (gRNA) that undergoes base pairing with the homologous DNA sequence. Pairing these various tools with transgene delivery methods offers a method to regulate the expression of specific genes in a defined cell population in the brains of awake, behaving rodents. These tools can be applied to a wide range of behavioural paradigms, electrophysiology experiments and biochemical analyses to understand the functional roles of a given gene in the brain.
