Fig. 2 |. Overview of CRISPRa/CRISPRi techniques.
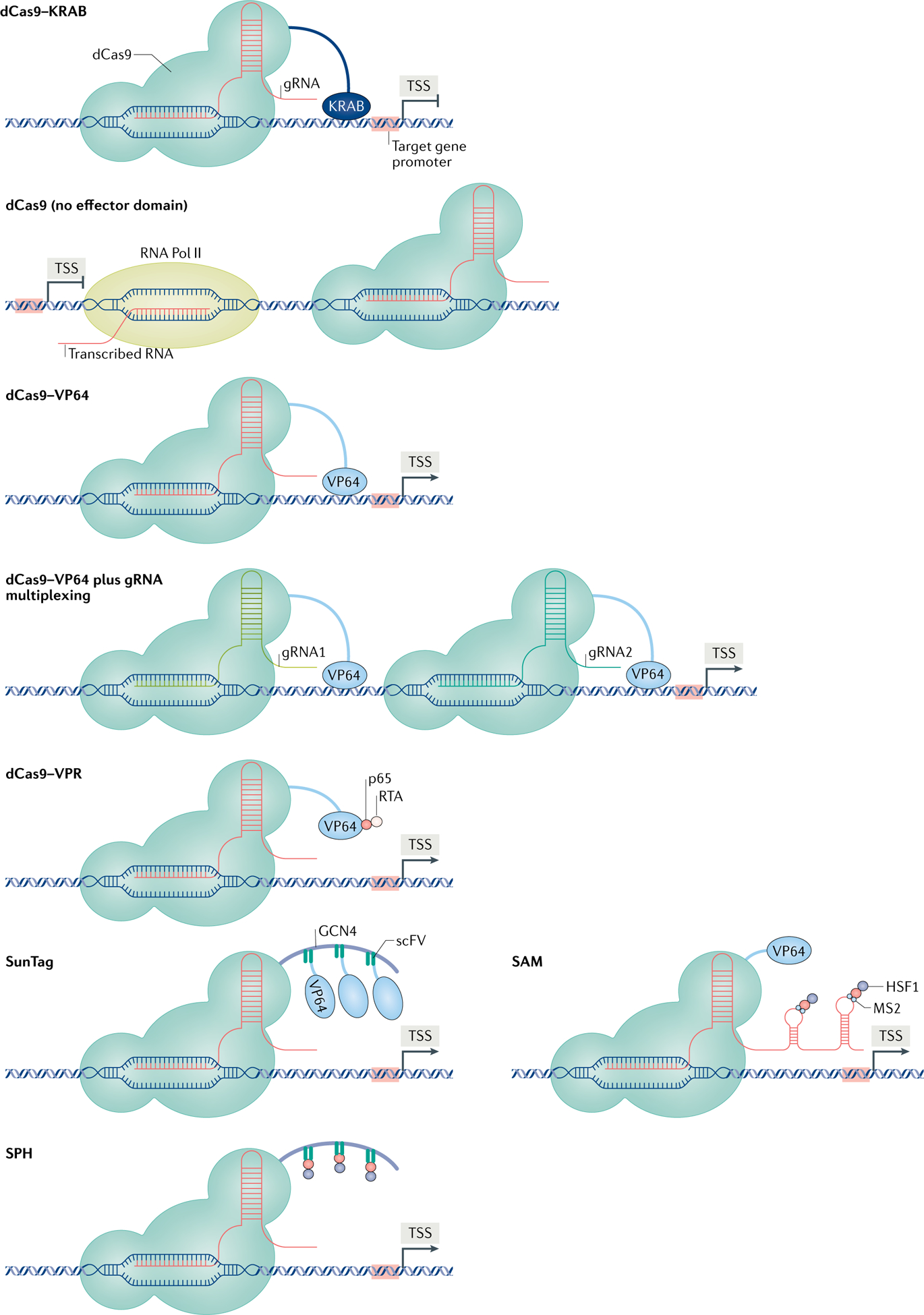
Clustered regularly interspaced short palindromic repeats (CRISPR) activation (CRISPRa)/CRISPR interference (CRISPRi) provides a powerful set of tools to study gene function through precise and modulatory regulation of gene expression. CRISPRi techniques include empty nuclease-dead CRISPR-associated protein 9 (dCas9) (that is, with no effector domain), which is thought to attenuate gene expression via steric hindrance of RNA polymerase II (Pol II) binding to DNA102, and dCas9 tethered to the Krüppel-associated box (KRAB) domain, a potent transcriptional repressor. The CRISPRa techniques were developed using a fusion between dCas9 and VP64, a potent viral transcriptional activator. This toolbox continues to expand to include new methods to increase effector recruitment to a single gene and increase the magnitude of gene induction. Combining dCas9–VP64 with multiplexed guide RNAs (gRNAs) directed towards several locations along the same promoter synergistically activates transcription45. In addition, dCas9 fused to multiple transcriptional effectors, including VP64, p65 and RTA (dCas9–VPR), substantially increases the magnitude of gene induction12,13. Synergistic activation mediator (SAM) combines dCas9–VP64 with a gRNA scaffold that selectively recruits an RNA-binding protein tethered to the p65 and HSF1 activation domains. Last, SunTag and SunTag–p65–HSF1 (SPH) use a dCas9-bound protein scaffold that acts as a binding site for single-chain variable fragment (scFv)-fused transcriptional activators. The effector domains for SunTag and SPH are VP64 and p65–HSF1, respectively. The CRISPRa/CRISPRi methods are illustrated on a spectrum showing their estimated repression and activation of gene expression. TSS, transcription start site.
