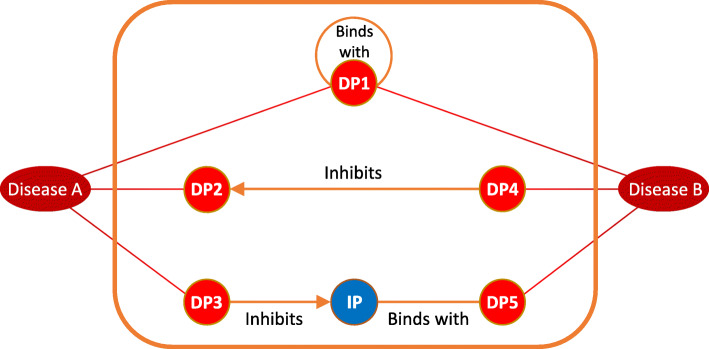
Fig. 1.
Schematic overview of the overlap, direct, and indirect scenarios that were extracted from the knowledge graph. Both diseases A and disease B have three disease proteins (DP) associated with them according to the manually curated subset of DisGeNet. DisGeNet describes that DP1 is known to be associated with both diseases, while the knowledge graph describes that it has a “binds with” relationship to itself. DP2 and DP4 have a direct “inhibits” relationship, and DP3 and DP5 are connected through an indirect path, by an intermediate protein (IP). The arrows between the proteins indicate which protein is the subject of the “inhibits” predicate, and which one its object. The “binds with” predicate was considered to be undirected by the experts, and therefore does not have a direction. Based on the paths in the knowledge graph, four feature sets are created, based on two methods to represent indirect paths, and both with and without the directional information of predicates