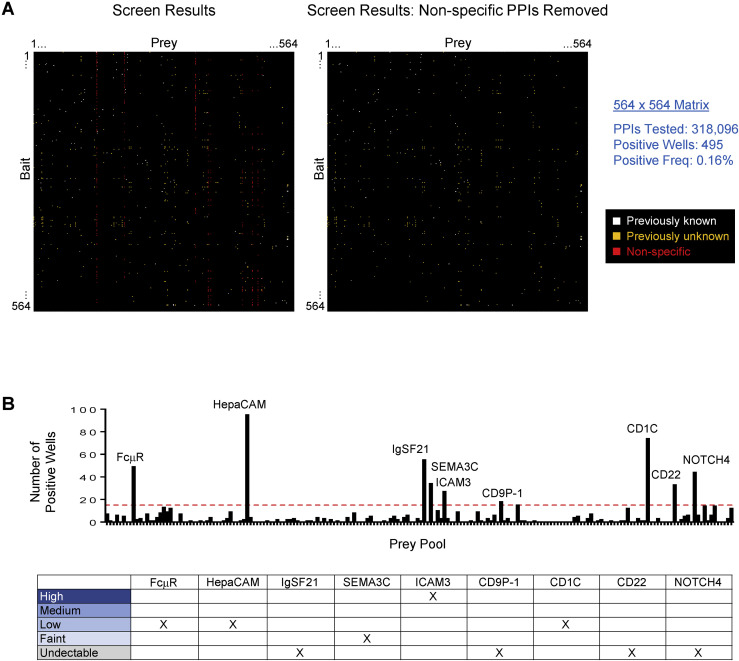
Figure S2.
Identification of Non-specific PPIs in Screen, Related to Data S3 and S4
(A) Left. Screen results for 564 × 564 matrix showing positive wells (≥2-fold over background). White, previously known PPI; orange, previously unknown PPI; red, non-specific PPI. Right. Screen results following removal of non-specific PPIs. Non-specific PPIs may result from misfolded proteins. Freq, frequency.
(B) Number of positive wells (≥2-fold over background) in the screen for each of the 188 prey pools. Prey pools with > 16 positive wells (above red dotted line) were classified as “sticky,” potentially exhibiting non-specific binding. In the screen, 16 PPIs for Siglec-10 were observed, many of which were PPIs with other Siglec subfamily members. As many Siglec-Siglec PPIs were observed in the screen, Siglec-10’s 16 PPIs were determined to not be the result of “stickiness,” and 16 PPIs was selected as a conservative cut-off for non-specific “sticky” prey to minimize the possibility of including false positives in the PPI dataset. Each prey in the screen was individually tested against mCherry-Fc bait and mock controls (data not shown) and positive binding signals were used to identify the “sticky” prey in each pool which were excluded from the PPI dataset. Table shows the protein expression level of the nine “sticky” prey in the conditioned media used for the screen. Qualitative expression levels were assessed by western blot.