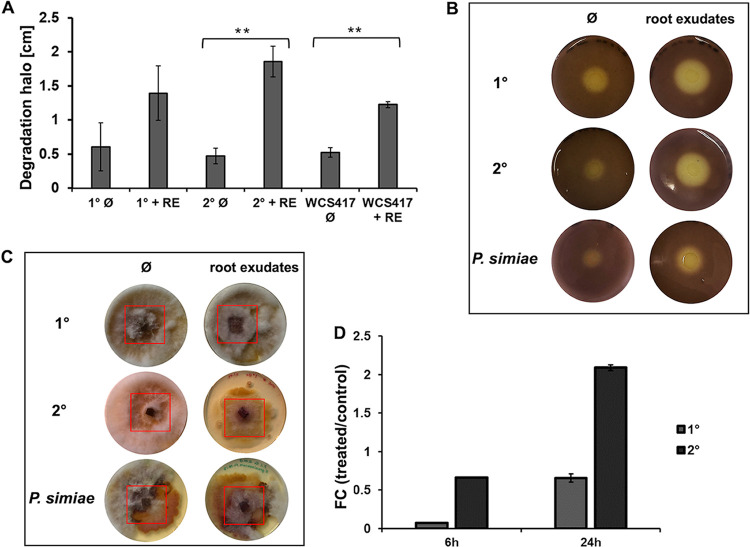
FIG 3.
Plant root exudates influence the chitin degradation capacity and fungal growth inhibition of P. luminescens 2° cells. (A) Chitin degradation halos (shown in panel B) in centimeters obtained during the chitin degradation assay. The plot shows the degradation halo (Ø) measured with ImageJ represented by the average and the standard deviation of three biological replicates (**, P ≤ 0.05). RE, root exudates. (B) Chitin degradation halo of P. luminescens 1° cells (1°), P. luminescens 2° cells (2°), and P. simiae WCS417 cultivated with or without root exudates. (C) Fungal growth inhibition assay using phytopathogenic Fusarium graminearum HM6PIS performed on YMG agar plates. P. luminescens 1° cells, P. luminescens 2° cells, and P. simiae WCS417, cultivated with and without plant root exudates, were placed around HM6PIS (red square) and incubated for 14 days at 26°C. (D) Expression level (fold change) of the chitinase-encoding gene (PluDJC_11885) in P. luminescens 1° and 2° cells using real-time qPCR analysis. P. luminescens 1° and 2° cells were cultivated in LB medium with root exudates (treated) or without (control) and collected at 6 h and 24 h postinoculation. Error bars represent standard deviation of at least three independently performed biological experiments. All pictures represent one characteristic of at least three independently performed experiments with similar outcomes.