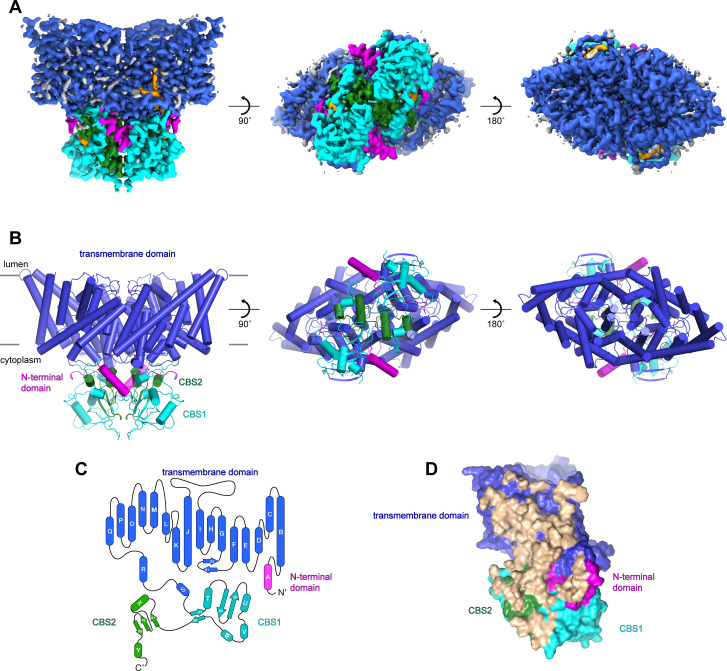
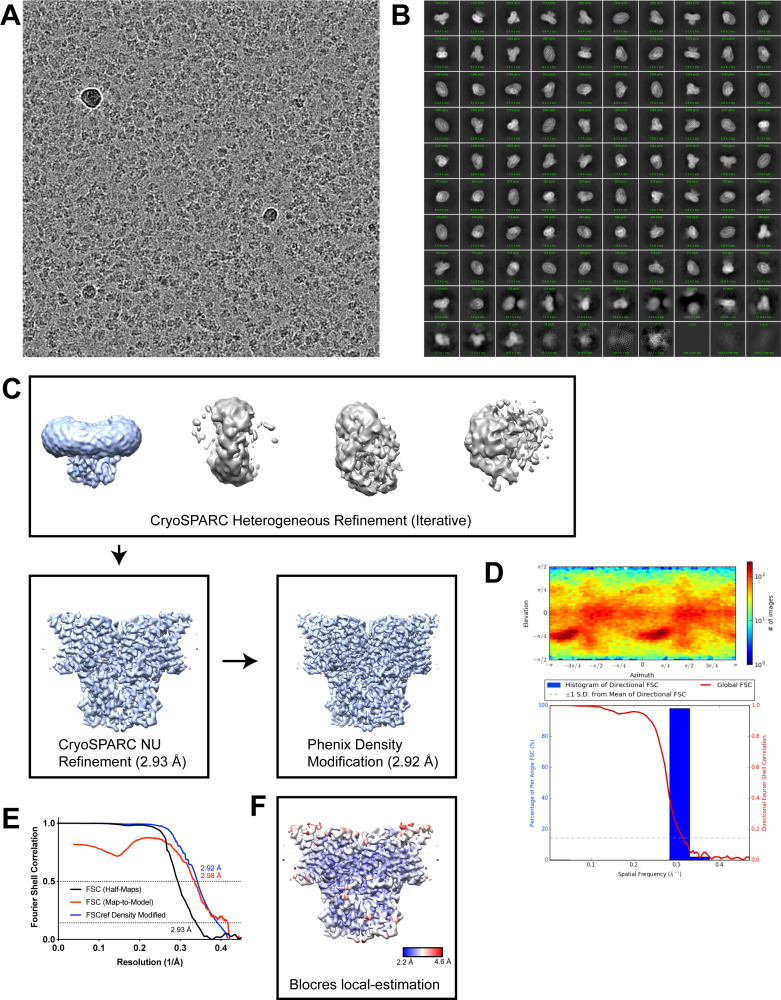
Figure 1. Structure of chicken CLC-7.
(A–B) Cryo-EM density map (A) and structure (B) of ggCLC-7 viewed from within the membrane (left), the cytoplasm (middle) and the lysosomal lumen (right) colored by domain with N-terminal domain in magenta, transmembrane domain in blue, CBS1 in cyan and CBS2 in green. Modeled non-protein densities are colored orange and unmodeled non-protein densities are colored grey in A. (C) Domain topology of ggCLC-7 colored by domain as in A. (D) Dimer interface with interacting residues colored in wheat.