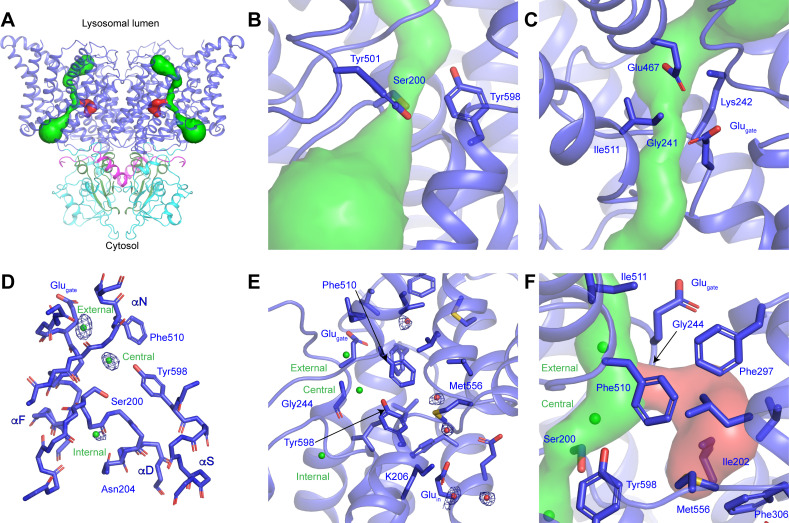
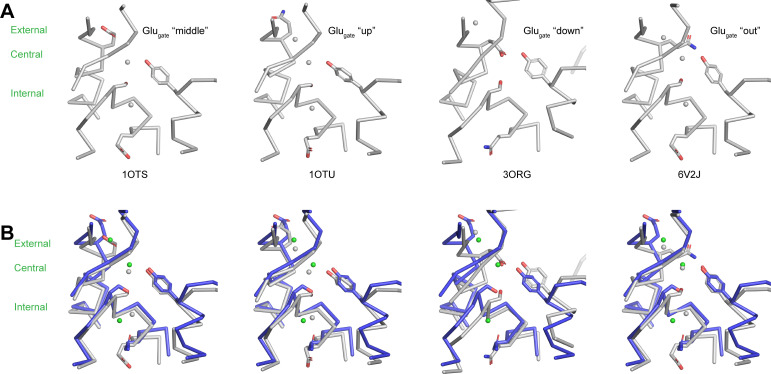
Figure 2. CLC-7 ion-conduction pathways.
(A) Each protomer of ggCLC-7 contains a Cl--conduction pathway, displayed as a green surface, and a putative H+-conduction pathway, displayed as a red surface. The N-terminal domain is colored in magenta, transmembrane domain in blue, CBS1 in cyan and CBS2 in green. (B) The cytosolic constriction of the Cl--conduction pathway formed by Ser200, Tyr501 and Tyr598 narrows the pathway to a minimum radius of 0.6 Å. (C) Two constrictions exist near the luminal entrance to the pathway formed by Glugate (Glu243), Lys242 and Ile511 and by Gly241, Lys242, Glu467. (D) Cl--binding sites (shown as green spheres) in the ggCLC-7 ion conduction pathway. Experimental cryo-EM density is shown as blue mesh countered at 10 σ threshold. Conserved residues are shown in sticks. (E) Ordered water molecules (shown as red spheres) are resolved in the hydrophobic gap between Glugate and Gluin and in the solvent-filled cavity in which Gluin resides. Experimental cryo-EM density is shown as blue mesh contoured at 7 σ threshold. (F) A potential H+-conduction pathway, shown as red surface, extends from near the central Cl--binding site through into the hydrophobic gap. The access from the Cl- pathway is lined by Gly244, Phe297 and Phe510. The pathway is separated from the cytosol by a constriction formed by Ile202, Phe306 and Met556.