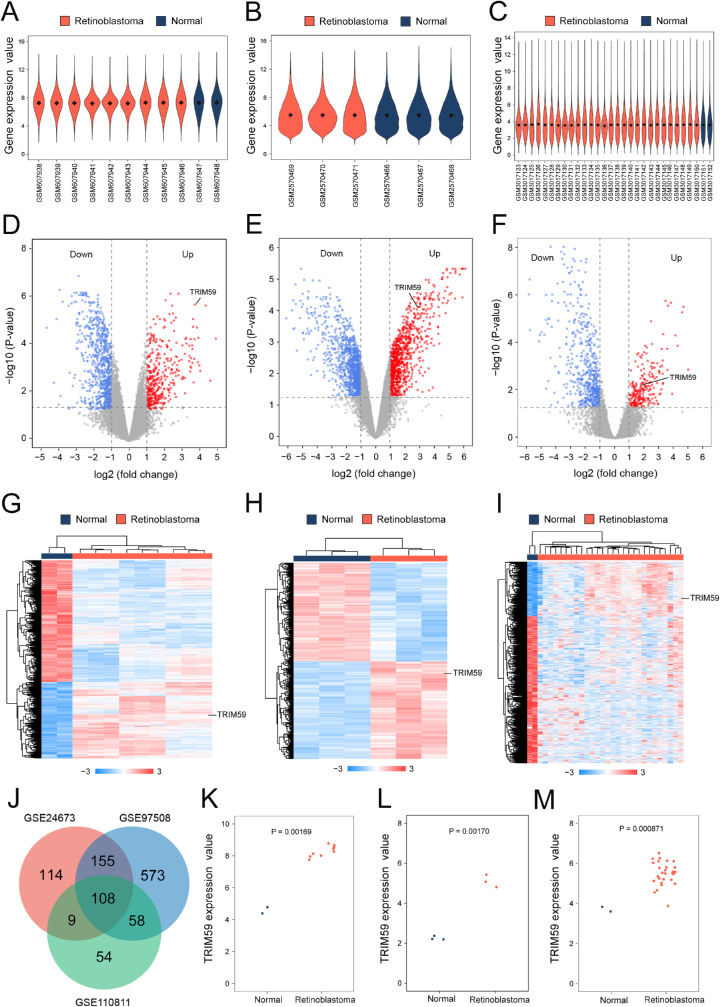
Figure 1.
Bioinformatic analysis of TRIM59 expression in retinoblastoma samples from the GEO database. Violin plot of the number of genes in retinoblastoma patient and normal control samples from the GSE24673 (A), GSE97508 (B), and GSE110811 (C) datasets. Black dots represent mean values of gene expression after sample normalization. Volcano plot of the distribution of DEGs in the GSE24673 (D), GSE97508 (E), and GSE110811 (F) datasets, mapping upregulated genes (red dots) and downregulated genes (blue dots). Genes showing no significant changes are marked as gray dots. Heatmap using hierarchical clustering of DEGs in the GSE24673 (G), GSE97508 (H), and GSE110811 (I) datasets. (J) Venn diagram DEGs were selected with a fold change > 2 and P < 0.01 in the GSE24673, GSE97508, and GSE110811 datasets. All three datasets showed overlap of 243 genes. Scatterplot of TRIM59 expression among retinoblastoma patient samples and normal control samples in the GSE24673 (K), GSE97508 (L), and GSE110811 (M) datasets.