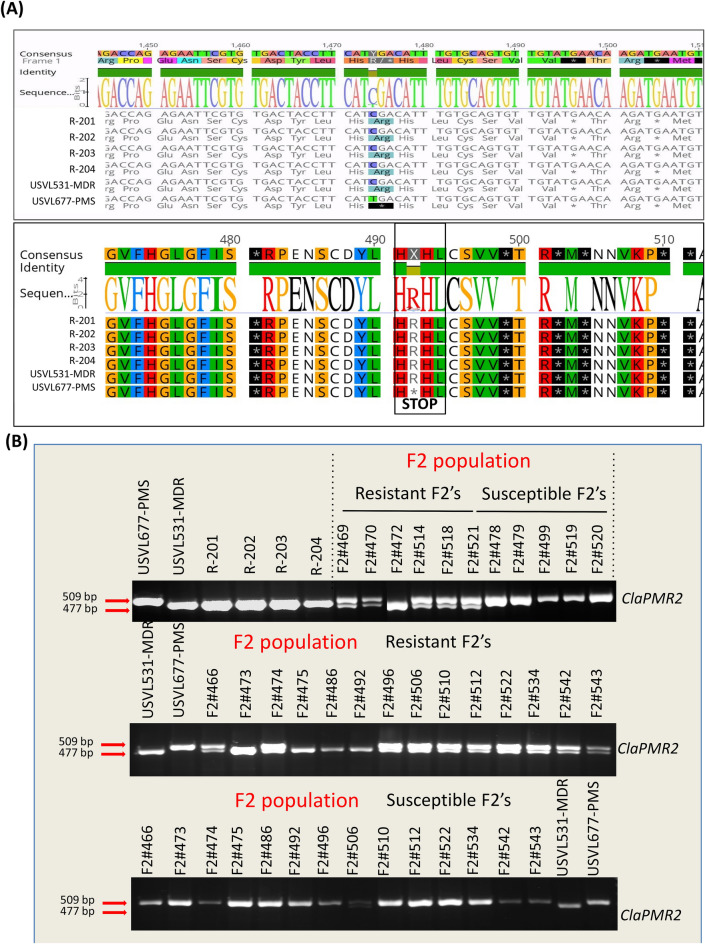
Figure 12.
SNPs derived CAPS validation on Parent, F2 and RIL introgressed watermelon lines using DNA Agarose gel. (A) Comparative nucleotide and amino acid consensus sequences with predicted secondary structure alignment of ClaPMR2 encoding protein in Parent (USVL531-MDR & USVL677-PMS), and RIL introgressed watermelon lines (R-201, R-202, R-203, R-204). Black line rectangle area HRH (C-T; Arg-STOP), represents the location of SNP region with substitution position Arg-STOP codon in (ClaPMR2, Citrullus lanatus PM Resistance gene). (B) DNA gel electrophoresis showing the CAPS marker analysis on ClaPMR2 gene in parent lines: USVL531-MDR & USVL677-PMS; RILs: R-201, R-202, R-203 and R-204 and F2 populations. Since the resistant phenotype is dominant, we observed ClaPMR2 loci, cosegregated with the resistant locus in heterozygous and homozygous individuals with PM resistance lines.