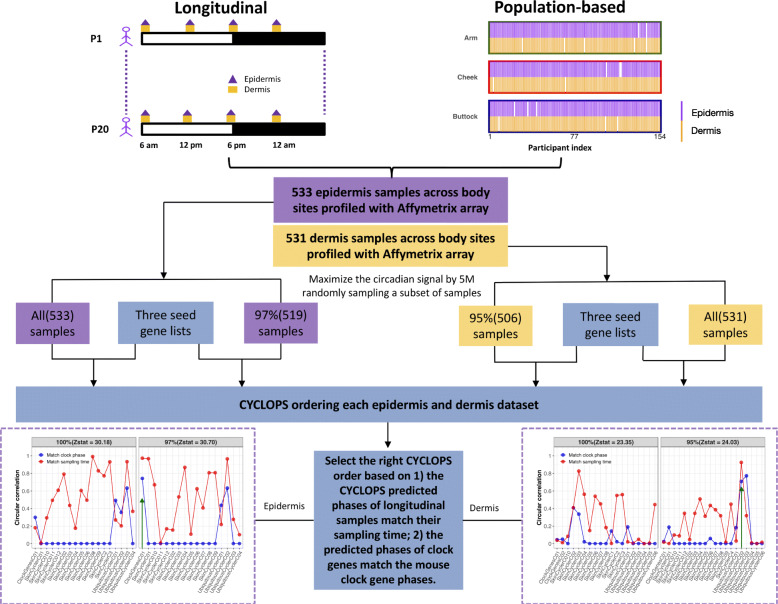
Fig. 2.
The pipeline for CYCLOPS ordering in epidermis and dermis samples. Each skin layer has two datasets. One dataset includes all epidermis (dermis) samples by combining longitudinal samples collected from 20 participants and population-based samples collected from 154 participants at three body sites (arm, cheek, and buttock). Another dataset has the maximum circadian signal in 5 million random subsetting 97% (95%) samples from all epidermis (dermis) samples. For CYCLOPS ordering these four datasets, we used three seed circadian gene lists, including (1) skin circadian genes, (2) human homologs of mouse ubiquitous circadian genes, and (3) clock and clock-associated genes (Additional file 4: Table S5). The high-quality CYCLOPS order should match both sampling time and clock phase. The matchability of sampling time and clock phase was measured by the corresponding circular correlation value. If fewer than 8 clock genes satisfied the cutoff (P < 0.01, fitmean > 16, rAMP > 0.1, and rsq > 0.1), the circular correlation value for “match clock phase” was set as 0. The optimal CYCLOPS order was indicated with a green arrow, with both high circular correlation values of sampling time and clock phase